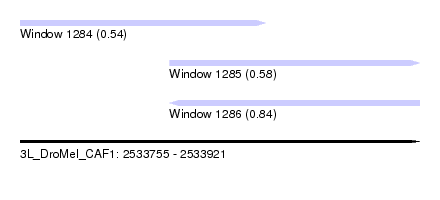
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,533,755 – 2,533,921 |
| Length | 166 |
| Max. P | 0.844292 |

| Location | 2,533,755 – 2,533,857 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2533755 102 + 23771897 AAAAACUUGUUAAUUAAGAAAUUAACAACGUUGCAUUACAUUAUGUUAACAUAUAGUGUGGGAAAUAUAGUGAAACAUUUCCACUCUUCUGCCUUAUAAUUC ...((((((((((((....))))))))).)))(((.((((((((((....))))))))))((((((..........)))))).......))).......... ( -20.20) >DroSec_CAF1 156607 82 + 1 AAAAACUUGUUAAUUAAGAAAUUAACAACGUUGCAUUACAUUAUGUUAACAUACAGUGUGGGAAA---UGUG-----------------UGCCUUAUAAUUC ...((((((((((((....))))))))).)))((((.(((((...(..((((...))))..).))---))))-----------------))).......... ( -14.10) >DroSim_CAF1 154176 84 + 1 AAAAACUUGUUAAUUAAGAAAUUAACAACGUUGCAUUACAUUAUGUUAACAUACAGUGUGGGAAU-ACAGUG-----------------UGCCUUAUAAUUC ...((((((((((((....))))))))).))).......((((((....(((((.((((....))-)).)))-----------------))...)))))).. ( -15.70) >consensus AAAAACUUGUUAAUUAAGAAAUUAACAACGUUGCAUUACAUUAUGUUAACAUACAGUGUGGGAAA_A_AGUG_________________UGCCUUAUAAUUC ...((((((((((((....))))))))).)))...((((((((((....))))..))))))......................................... (-11.90 = -11.90 + 0.00)

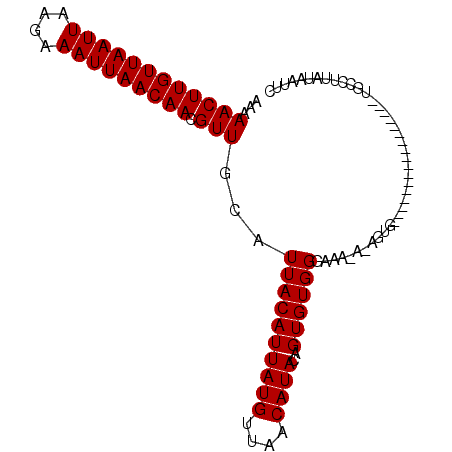
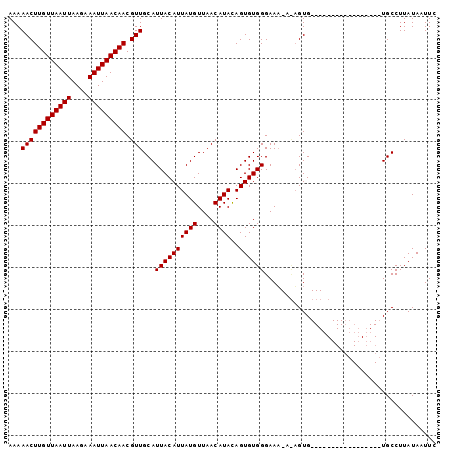
| Location | 2,533,817 – 2,533,921 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -17.65 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2533817 104 + 23771897 AAAUAUAGUGAAACAUUUCCACUCUUCUGCCUUAUAAUUCCCAACUAGCCAUUUCGUUUGACUUAUUAAGGGAAGUAAUCCUCUUUUGUUCAGAGGGUUAAACG ......((((.........))))((((..((((((((.((..(((..........))).)).))).)))))))))(((((((((.......))))))))).... ( -17.10) >DroSec_CAF1 156669 84 + 1 AAA---UGUG-----------------UGCCUUAUAAUUCCCAACUAGCCAUUUCGUUCGACUUAUUAAGGGAAGUAAUCCUCUGUUGUUCAGAGGGUUAAACG ...---....-----------------..........(((((...........................))))).((((((((((.....)))))))))).... ( -17.93) >DroSim_CAF1 154238 86 + 1 AAU-ACAGUG-----------------UGCCUUAUAAUUCCCAACUAGCCAUUUCGUUCGACUUAUUAAGGGAAGUAAUCCUCUGUUGUUCAGAGGGUUAAACG ...-......-----------------..........(((((...........................))))).((((((((((.....)))))))))).... ( -17.93) >consensus AAA_A_AGUG_________________UGCCUUAUAAUUCCCAACUAGCCAUUUCGUUCGACUUAUUAAGGGAAGUAAUCCUCUGUUGUUCAGAGGGUUAAACG .....................................(((((...........................))))).((((((((((.....)))))))))).... (-16.40 = -16.73 + 0.33)

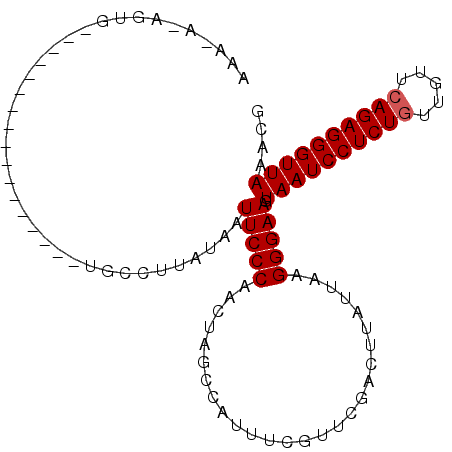
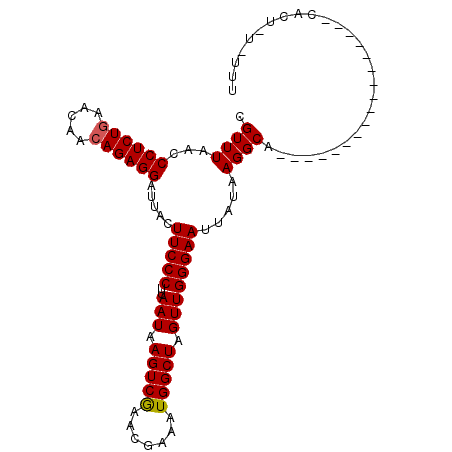
| Location | 2,533,817 – 2,533,921 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2533817 104 - 23771897 CGUUUAACCCUCUGAACAAAAGAGGAUUACUUCCCUUAAUAAGUCAAACGAAAUGGCUAGUUGGGAAUUAUAAGGCAGAAGAGUGGAAAUGUUUCACUAUAUUU .((((...(((((.......))))).....(((((..(((.(((((.......))))).)))))))).....)))).....((((((.....))))))...... ( -22.80) >DroSec_CAF1 156669 84 - 1 CGUUUAACCCUCUGAACAACAGAGGAUUACUUCCCUUAAUAAGUCGAACGAAAUGGCUAGUUGGGAAUUAUAAGGCA-----------------CACA---UUU .((((...((((((.....)))))).....(((((..(((.(((((.......))))).)))))))).....)))).-----------------....---... ( -19.20) >DroSim_CAF1 154238 86 - 1 CGUUUAACCCUCUGAACAACAGAGGAUUACUUCCCUUAAUAAGUCGAACGAAAUGGCUAGUUGGGAAUUAUAAGGCA-----------------CACUGU-AUU .((((...((((((.....)))))).....(((((..(((.(((((.......))))).)))))))).....)))).-----------------......-... ( -19.20) >consensus CGUUUAACCCUCUGAACAACAGAGGAUUACUUCCCUUAAUAAGUCGAACGAAAUGGCUAGUUGGGAAUUAUAAGGCA_________________CACU_U_UUU .((((...((((((.....)))))).....(((((..(((.(((((.......))))).)))))))).....))))............................ (-18.42 = -18.53 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:22 2006