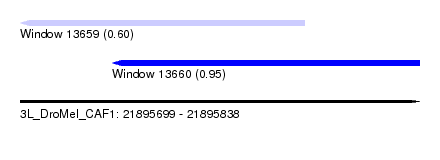
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,895,699 – 21,895,838 |
| Length | 139 |
| Max. P | 0.953135 |

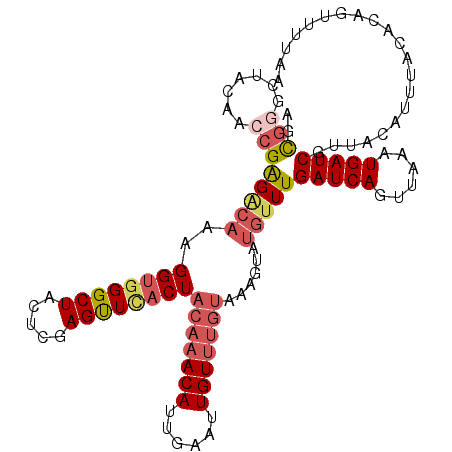
| Location | 21,895,699 – 21,895,798 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21895699 99 - 23771897 ACUACAAACAUUGAAUUGUUUGUAAAGUAUGUUUGAUCAGUUAAAUGAUCGGAACAUUUACGCAGUU------------CGUCAUGAACAGAUUACACUGGUGUCAUUAUU ............(((((((..(((((.....((((((((......))))))))...)))))))))))------------)...((((.(((......)))...)))).... ( -22.20) >DroSec_CAF1 16385 108 - 1 ACUACAAACAUUGAAUUGUUUGUAAAGUGUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUUGAAGUGCUUCUCGCCAUGAACAGAUUACACUGGUGUCA---UU ..((((((((......))))))))(((((((..((((((......)))))).)))))))(((((((.(((..((.(((.......)))))..))).))).))))..---.. ( -26.20) >DroSim_CAF1 16557 108 - 1 ACUACAAACAUUGAAUUGUUUGUAAAGUGUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUUAAAGUGCUUCUCGCCAUGAACAGAUUACACUGGUGUCA---UU ..((((((((......))))))))(((((((..((((((......)))))).)))))))(((((((.(((..((.(((.......)))))..))).))).))))..---.. ( -26.10) >consensus ACUACAAACAUUGAAUUGUUUGUAAAGUGUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUU_AAGUGCUUCUCGCCAUGAACAGAUUACACUGGUGUCA___UU ..((((((((......))))))))((((((...((((((......))))))..)))))).........................(((.(((......)))...)))..... (-19.17 = -19.50 + 0.33)

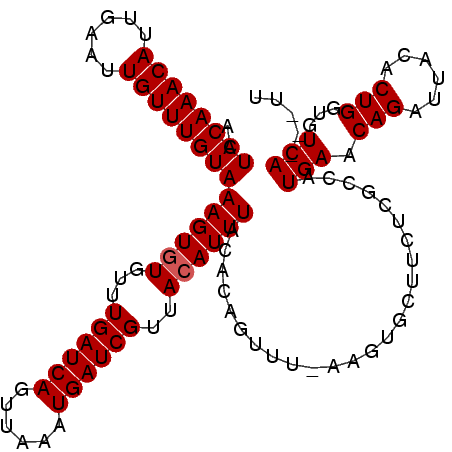
| Location | 21,895,731 – 21,895,838 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -17.09 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21895731 107 - 23771897 UGCGGGCUACAACCGAGGCAAAGGUGGGCUACUCGAGUUGACUACAAACAUUGAAUUGUUUGUAAAGUAUGUUUGAUCAGUUAAAUGAUCGGAACAUUUACGCAGUU---- ((((.(((.((((((((((........))..)))).))))..((((((((......)))))))).)))(((((((((((......))))).))))))...))))...---- ( -32.60) >DroSec_CAF1 16422 111 - 1 AGCGGGCUACAACCGAGACAAUGGUGGGCUACUCGAGUUCACUACAAACAUUGAAUUGUUUGUAAAGUGUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUUGAA ..(((.......)))((((..(((((((((.....)))))))((((((((......))))))))(((((((..((((((......)))))).))))))).))..))))... ( -31.30) >DroSim_CAF1 16594 111 - 1 AGCGGGCUACAACCGAGACAAUGGUGGGCUACUCGAGUUCACUACAAACAUUGAAUUGUUUGUAAAGUGUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUUAAA ..(((.......)))((((..(((((((((.....)))))))((((((((......))))))))(((((((..((((((......)))))).))))))).))..))))... ( -31.30) >DroEre_CAF1 14607 88 - 1 AGCGAUCUACACCCGGGACAAGGGUGGGCUACUCGAGUUUUCUACA-ACAUGGAAUUGU-------------UUGAUCAGUUCAAUGAUCAUUACA---------UUUUAA ...((((..(((((.......)))))((((..(((((..(((((..-...)))))...)-------------))))..))))....))))......---------...... ( -19.70) >DroYak_CAF1 20052 110 - 1 AGUCAACUACACAAGAGACACAGGUGGGCUACUCGAGCUUACUACA-ACAUUGAAUUGUUUGUGAAGUAUGUUUGAUCAGUUCAAUGAUCAUUACAUGCACGACCUUUUAA .(((..............((((((((((((.....))))))))..(-(((......))))))))..((((((.((((((......))))))..))))))..)))....... ( -30.50) >consensus AGCGGGCUACAACCGAGACAAAGGUGGGCUACUCGAGUUCACUACAAACAUUGAAUUGUUUGUAAAGUAUGUUUGAUCAGUUAAAUGAUCGUUACAUUUACACAGUUUUAA ..(((.......)))(((((..((((((((.....))))))))(((((((......)))))))......)))))(((((......)))))..................... (-17.09 = -18.78 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:11 2006