
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,852,138 – 21,852,243 |
| Length | 105 |
| Max. P | 0.626567 |

| Location | 21,852,138 – 21,852,243 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -18.44 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21852138 105 + 23771897 -----------CACACAACCGUAAUGAUCAAUCA----AUCAAUCGAACAUAGACCUGUCCAGUAUUUCCACUUGCCCUGGCUACCGGCGGCCUACAUGCUGGUAUCUCAGGCUUGGCGG -----------.......((((..((((......----))))......((.((.((((.(((((((........(((((((...)))).)))....))))))).....)))))))))))) ( -27.30) >DroPse_CAF1 71803 107 + 1 -----------CACACAAA--UACAAAUGAAAAAAUACAACAAUAAAACGUAGACCUGUCCAGUAUUUCCACUUGCCCUGGCUACCGGCGGCCUACAUGCUGGUAUUUCAGGCUUAGCGG -----------........--...........................(((.((((((.(((((((........(((((((...)))).)))....))))))).....)))).)).))). ( -23.60) >DroSim_CAF1 31717 109 + 1 -----------CACACAACCGUAAUGAUCAAUCAAUCAAUCAAUCGAACAUAGACCUGUCCAGUAUUUCCACUUGCCCUGGCUACCGGCGGCCUACAUGCUGGUAUCUCAGGCUUGGCGG -----------.......((((..((((......))))..........((.((.((((.(((((((........(((((((...)))).)))....))))))).....)))))))))))) ( -27.30) >DroEre_CAF1 32563 105 + 1 -----------CACAAAACCGAAAUGAUCAAUCA----AUCGAACGAACAUAGACCUGUCCAGUAUUUCCACUUGCCCUGGCUACCGGCGGCCUACAUGCUGGUAUCUCAGGCUUGGCGG -----------.......(((...((((......----))))......((.((.((((.(((((((........(((((((...)))).)))....))))))).....)))))))).))) ( -24.80) >DroWil_CAF1 18288 118 + 1 AUCUUCAAAAAAAAAAAAACAAAAAAAACAAAUAAUU-ACCAAUCAUA-UUAGACCUGCCCAGUAUUUCCACUGGCUUUGGCUACCGGUGGCCUACAUGCUGGUAUUUCAGGCCUUACGG .....................................-..........-..((.((((.(((((((..(((((((.........))))))).....))))))).....)))).))..... ( -21.80) >DroMoj_CAF1 87836 113 + 1 AUAUCGAAAAAAAAAAAAUCGAAUUAAAAAA---AUG---UAAUACUA-UUAGACCUGUCCAGUAUUUCCACUUGCUUUGGCUACCGGUGGUCUACAUGCUGGUAUUUCAGGCUUAGCGG ...((((...........)))).........---...---.....(..-(((..((((.(((((((..(((((.((....))....))))).....))))))).....))))..)))..) ( -23.70) >consensus ___________CACAAAACCGAAAUAAUCAAUCAAU__AUCAAUCGAACAUAGACCUGUCCAGUAUUUCCACUUGCCCUGGCUACCGGCGGCCUACAUGCUGGUAUCUCAGGCUUGGCGG ..................................................(((.((((.(((((((........(((((((...)))).)))....))))))).....)))).))).... (-18.44 = -17.72 + -0.72)

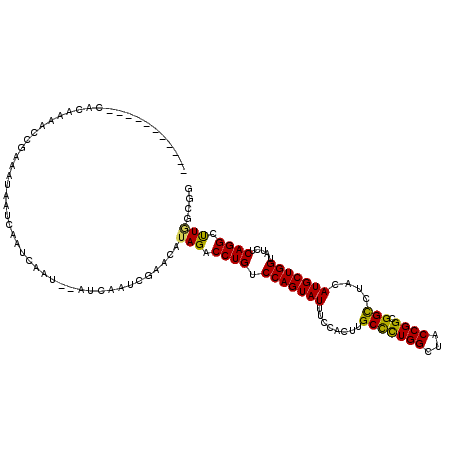
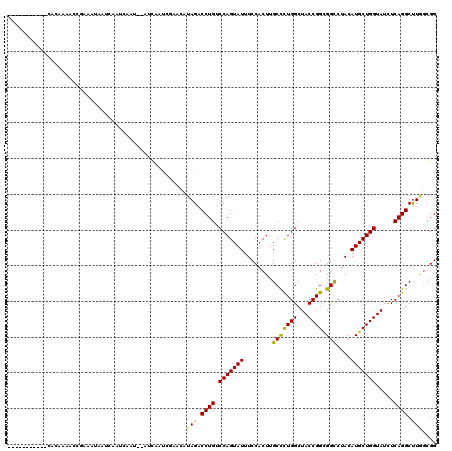
| Location | 21,852,138 – 21,852,243 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -22.09 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21852138 105 - 23771897 CCGCCAAGCCUGAGAUACCAGCAUGUAGGCCGCCGGUAGCCAGGGCAAGUGGAAAUACUGGACAGGUCUAUGUUCGAUUGAU----UGAUUGAUCAUUACGGUUGUGUG----------- .(((..((((..........(.(((((((((.((((((.(((.......)))...))))))...))))))))).)(((..(.----...)..))).....))))..)))----------- ( -30.90) >DroPse_CAF1 71803 107 - 1 CCGCUAAGCCUGAAAUACCAGCAUGUAGGCCGCCGGUAGCCAGGGCAAGUGGAAAUACUGGACAGGUCUACGUUUUAUUGUUGUAUUUUUUCAUUUGUA--UUUGUGUG----------- ..((.((....(((((((.((((((((((((.((((((.(((.......)))...))))))...))))))))......)))))))))))....)).)).--........----------- ( -29.30) >DroSim_CAF1 31717 109 - 1 CCGCCAAGCCUGAGAUACCAGCAUGUAGGCCGCCGGUAGCCAGGGCAAGUGGAAAUACUGGACAGGUCUAUGUUCGAUUGAUUGAUUGAUUGAUCAUUACGGUUGUGUG----------- .(((..((((..........(.(((((((((.((((((.(((.......)))...))))))...))))))))).)(((..(((....)))..))).....))))..)))----------- ( -33.70) >DroEre_CAF1 32563 105 - 1 CCGCCAAGCCUGAGAUACCAGCAUGUAGGCCGCCGGUAGCCAGGGCAAGUGGAAAUACUGGACAGGUCUAUGUUCGUUCGAU----UGAUUGAUCAUUUCGGUUUUGUG----------- .(((.(((((.(((((....(.(((((((((.((((((.(((.......)))...))))))...))))))))).)....(((----(....)))))))))))))).)))----------- ( -34.50) >DroWil_CAF1 18288 118 - 1 CCGUAAGGCCUGAAAUACCAGCAUGUAGGCCACCGGUAGCCAAAGCCAGUGGAAAUACUGGGCAGGUCUAA-UAUGAUUGGU-AAUUAUUUGUUUUUUUUGUUUUUUUUUUUUUGAAGAU ((....)).......((((((((((((((((.((((((.(((.......)))...))))))...))))).)-)))).)))))-)................((((((........)))))) ( -31.30) >DroMoj_CAF1 87836 113 - 1 CCGCUAAGCCUGAAAUACCAGCAUGUAGACCACCGGUAGCCAAAGCAAGUGGAAAUACUGGACAGGUCUAA-UAGUAUUA---CAU---UUUUUUAAUUCGAUUUUUUUUUUUUCGAUAU ..((((.((.((......))))...((((((.((((((.(((.......)))...))))))...)))))).-))))....---...---.........((((...........))))... ( -24.20) >consensus CCGCCAAGCCUGAAAUACCAGCAUGUAGGCCGCCGGUAGCCAGGGCAAGUGGAAAUACUGGACAGGUCUAUGUUCGAUUGAU__AUUGAUUGAUCAUUACGGUUGUGUG___________ .......((.((......))))(((((((((.((((((.(((.......)))...))))))...)))))))))............................................... (-22.09 = -22.53 + 0.45)

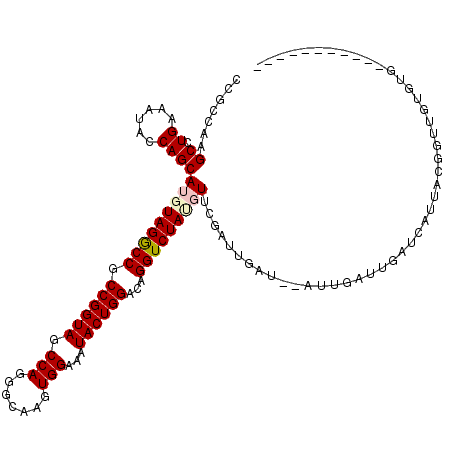
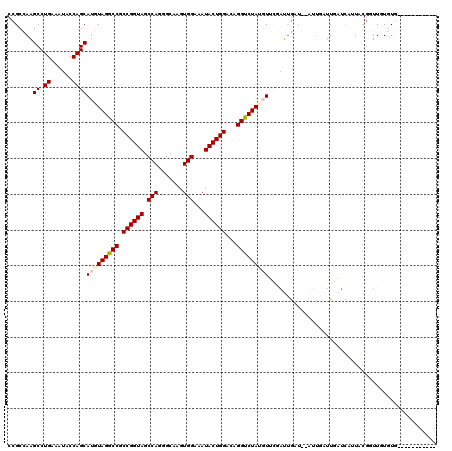
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:02 2006