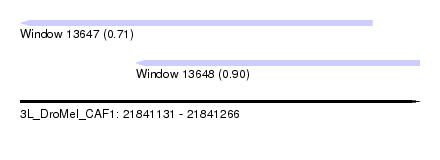
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,841,131 – 21,841,266 |
| Length | 135 |
| Max. P | 0.898304 |

| Location | 21,841,131 – 21,841,250 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.30 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21841131 119 - 23771897 GUUGAGCAAGAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUCUGUUGGGUGCCUAAGCUGCAAAGAGACAGAAAAUUAUUGAAACGAUUAGUUUGGCACGAAUAAAUCAUUU (((((((((.(....).)))))))))..(((((((....))))))).((((((...(((.......)))....)))))).............((((.(((((...))))).)))).... ( -28.30) >DroSec_CAF1 22055 119 - 1 GUUGAGCAAGAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUACAUUUGUUGGGUGCCUAAGCUGCAAAGAGACAGAAGAGUAUUGAAACGGUUAGUUUGGCACGCAUAAAUCAUUU (((((((((.(....).)))))))))...((((((....)))))).((((((.(.(((((.(((((((...((.((......)).))......).))))))))))).)))))))..... ( -29.50) >DroSim_CAF1 20743 119 - 1 GUUGAGCAAGAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUUUGUUGGGUGCCUAAGCUGCAAAGAGAGAGAAGAUUAUUGAAACGGUUAGUUUGGCACGCAUAAAUCAUUU (((((((((.(....).)))))))))..(((((((....)))))))((((((.(.(((((.(((((((.......((........))......).))))))))))).)))))))..... ( -30.32) >DroEre_CAF1 20789 118 - 1 GUUGAGCAAAAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUUUGUUGGGCGUCUAAGCUGCAAGGAGACAGAAGAA-AUGGAAACGGUUAGUUUGGCACGCAUUAAUCAUUU (((((((((........)))))))))..(((((((....)))))))....((((((((((((((((((..(....).......-.((....))).)))))))).)))).)))))..... ( -32.50) >DroYak_CAF1 17625 118 - 1 GUUGAGCAAAAAUACUGUUGCUUAGCUUAUGCAUUUAGAGAUGUAUAUUUGUUGGGUGCCUAAGCUGAAAUCAGACAGAAGAA-AUUGAAACGGUUAGUGAGGCACGCAUAAAUCAUUU (((((((((........))))))))).((((((((....))))))))(((((.(.((((((.(((((...((((.........-.))))..)))))....)))))).))))))...... ( -31.60) >consensus GUUGAGCAAGAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUUUGUUGGGUGCCUAAGCUGCAAAGAGACAGAAGAAUAUUGAAACGGUUAGUUUGGCACGCAUAAAUCAUUU (((((((((........)))))))))..(((((((....)))))))......((.(((((....(((........)))..........((((.....))))))))).)).......... (-23.26 = -24.30 + 1.04)


| Location | 21,841,170 – 21,841,266 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.56 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.98 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21841170 96 - 23771897 UUUUCGUAUA-UUUAUUGUUGAGCAAGAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUCUGUUGGGUGCCUAAGCUGCAAAGAGACAGAA ..........-......(((((((((.(....).)))))))))..(((((((....))))))).((((((...(((.......)))....)))))). ( -24.30) >DroVir_CAF1 17440 74 - 1 UAU-----UAUUUUAAUGUUGAGCAAAAAUACUGUUGCUUAGCGUUUGUAUUUAGA----------CUUUG--------CACUGCAAAGAGACAAAA ...-----(((...((((((((((((........)))))))))))).)))......----------(((((--------(...))))))........ ( -17.90) >DroGri_CAF1 73754 76 - 1 UAAU--AUUAUUUUAAUGUUGAGCGAAAAUACUGUUGCUUAGCGGUAGUAUUUAGU----------UGUUG--------CACUGCAAAGAGAC-AAA .(((--(((((.....((((((((((........))))))))))))))))))..((----------(.(((--------(...))))...)))-... ( -16.70) >DroEre_CAF1 20827 96 - 1 UUUUCGUAUA-AUUAUUGUUGAGCAAAAAUACUGUUGCUUAGCUGAUGCAUUUAGAGAUGUAUAUUUGUUGGGCGUCUAAGCUGCAAGGAGACAGAA (((((.....-......(((((((((........)))))))))...((((.....((((((.((.....)).))))))....)))).)))))..... ( -21.40) >DroYak_CAF1 17663 96 - 1 UUUUUGUAUA-UUUAUUGUUGAGCAAAAAUACUGUUGCUUAGCUUAUGCAUUUAGAGAUGUAUAUUUGUUGGGUGCCUAAGCUGAAAUCAGACAGAA (((((((.((-........)).)))))))..(((((..((((((((.((((((((((((....)))).)))))))).)))))))).....))))).. ( -24.00) >DroMoj_CAF1 69800 78 - 1 UAUUUGUUUAUGUUAAUGUUGAGCAAAAAUACUGUUGCUUAGCGUUUGUAUUUAGU----------UUUCG--------CACUGCAAAGAAA-AAGA ..(((((..(((..((((((((((((........))))))))))))..)))..(((----------.....--------.))))))))....-.... ( -14.50) >consensus UAUU_GUAUA_UUUAAUGUUGAGCAAAAAUACUGUUGCUUAGCGGAUGCAUUUAGA__________UGUUG________AACUGCAAAGAGACAAAA .................(((((((((........)))))))))...................................................... (-10.12 = -9.98 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:59 2006