
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,835,945 – 21,836,093 |
| Length | 148 |
| Max. P | 0.978162 |

| Location | 21,835,945 – 21,836,055 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21835945 110 + 23771897 UUCCCUGCCCUGUUUUAACUUUUUCAUUCUGUCUGGCAUCAGCACAUCUCAGCAUCCUUUAUCAUACAUCCUCGUGUGGAUGAUGAUGACGGUGAUGUGGACAUGGCUUU ......(((.((((..............(((........)))((((((.......((.(((((((.(((((......)))))))))))).)).)))))))))).)))... ( -31.20) >DroSec_CAF1 16824 110 + 1 UCCCCUGCCCUGUUUUAACUUUUUCAUUCUGCCUGGCAUCAGCACAUCUCAGCAUCCUUUAUCAUACAUCCUCGUGUGGAUGAUGAUGACGGUGAUGUGGACAUGGCUUU .....((((..((.................))..))))..(((.(((.((.((((((((((((((.(((((......)))))))))))).)).))))).)).)))))).. ( -31.13) >DroSim_CAF1 15488 109 + 1 UC-CCUGCCCUGUUUUAACUUUUUCAUUCUGCCUGGCAUCAGCACAUCUCAGCAUCCUUUAUCAUACAUCCUCGUGUGGAUGAUGAUGACGGUGAUGUGGACAUGGCUUU ..-..((((..((.................))..))))..(((.(((.((.((((((((((((((.(((((......)))))))))))).)).))))).)).)))))).. ( -31.13) >DroEre_CAF1 15671 99 + 1 UCC-CUGCCCUGUUUUAACUUUUUCAUUUGGCCUGGCAUCA----------GCAUCCUUUAUCAUCCAUCCUCGUGUGGAUGAUGAUGAAGACGAUGUGGACAUGGCUUU ...-..(((.(((((...............((...))....----------(((((((((((((((.((((......))))))))))))))..)))))))))).)))... ( -29.40) >consensus UCCCCUGCCCUGUUUUAACUUUUUCAUUCUGCCUGGCAUCAGCACAUCUCAGCAUCCUUUAUCAUACAUCCUCGUGUGGAUGAUGAUGACGGUGAUGUGGACAUGGCUUU ......(((.(((((.................(((....))).........((((((((((((((.(((((......)))))))))))).)).)))))))))).)))... (-25.90 = -26.40 + 0.50)



| Location | 21,835,945 – 21,836,055 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21835945 110 - 23771897 AAAGCCAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGUAUGAUAAAGGAUGCUGAGAUGUGCUGAUGCCAGACAGAAUGAAAAAGUUAAAACAGGGCAGGGAA ....((.(((((...(((.((((((((((((((......))))).)))))....)))).)))..(((.(((....))))))...................))))).)).. ( -29.20) >DroSec_CAF1 16824 110 - 1 AAAGCCAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGUAUGAUAAAGGAUGCUGAGAUGUGCUGAUGCCAGGCAGAAUGAAAAAGUUAAAACAGGGCAGGGGA ....((.(((((.((((......((((((((((......))))).)))))....))))(((...((.((....))))..)))..................))))).)).. ( -30.20) >DroSim_CAF1 15488 109 - 1 AAAGCCAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGUAUGAUAAAGGAUGCUGAGAUGUGCUGAUGCCAGGCAGAAUGAAAAAGUUAAAACAGGGCAGG-GA ....((.(((((.((((......((((((((((......))))).)))))....))))(((...((.((....))))..)))..................))))).)-). ( -30.00) >DroEre_CAF1 15671 99 - 1 AAAGCCAUGUCCACAUCGUCUUCAUCAUCAUCCACACGAGGAUGGAUGAUAAAGGAUGC----------UGAUGCCAGGCCAAAUGAAAAAGUUAAAACAGGGCAG-GGA ....((.(((((.((((..(((.((((((((((......)))).)))))).)))))))(----------((....)))......................))))).-)). ( -27.10) >consensus AAAGCCAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGUAUGAUAAAGGAUGCUGAGAUGUGCUGAUGCCAGGCAGAAUGAAAAAGUUAAAACAGGGCAGGGGA ...(((.((((..(((((((...((((((((((......))))).)))))...))..((........)))))))...))))..........((....))..)))...... (-22.91 = -23.10 + 0.19)

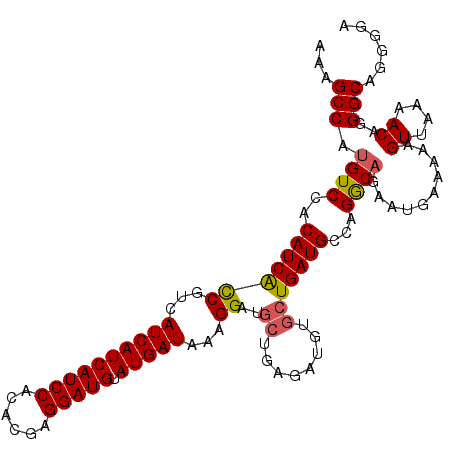
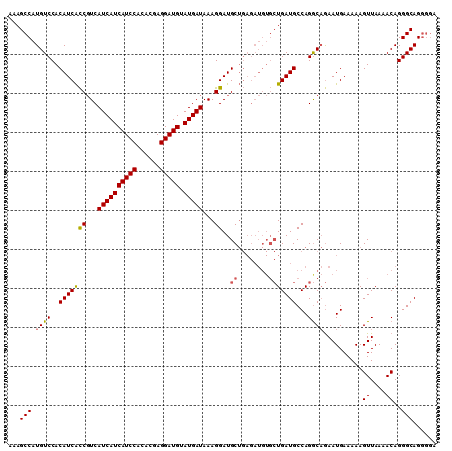
| Location | 21,835,978 – 21,836,093 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.34 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21835978 115 - 23771897 UGUGUCACAUAUAUCAUCAUUUUGUCUUCCUUUUUGACAAAGC-CAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGU-AUGAUAAAGGAUGCUGAGAUGUGCU-GAUGCCA .(((((((((((.(((.(((((((((.........))))))..-.............((...((((((((((......))))).-)))))...))))).))).))))).)-))))).. ( -31.40) >DroPse_CAF1 52624 92 - 1 CGUGUCACA----UCAUCGUUUUGUCCUUCUCUUUGACAAAACACAUG---------------UCAUCAUCUCGUGGGGCAUGCAAUGAUAAAGGAUGCUG-------CUGGCUGCCA ...((((((----.((((((((((((.........)))))))).((((---------------((.((((...))))))))))...........)))).))-------.))))..... ( -24.70) >DroSec_CAF1 16857 115 - 1 UGUGUCACAUAUAUCAUCGUUUUGUCUUCCUUUUUGACAAAGC-CAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGU-AUGAUAAAGGAUGCUGAGAUGUGCU-GAUGCCA .(((((((((((..(((.((((((((.........))))))))-.)))......(((.((((((((((((((......))))).-)))))....)))).))).))))).)-))))).. ( -34.30) >DroSim_CAF1 15520 115 - 1 UGUGUCACAUAUAUCAUCGUUUUGUCUUCCUUUUUGACAAAGC-CAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGU-AUGAUAAAGGAUGCUGAGAUGUGCU-GAUGCCA .(((((((((((..(((.((((((((.........))))))))-.)))......(((.((((((((((((((......))))).-)))))....)))).))).))))).)-))))).. ( -34.30) >DroEre_CAF1 15703 105 - 1 UGUGUCACAUAUAUCAUCGUUUUGUCUUCAUUUUUGACAAAGC-CAUGUCCACAUCGUCUUCAUCAUCAUCCACACGAGGAUGG-AUGAUAAAGGAUGC----------U-GAUGCCA .(((((((((....(((.((((((((.........))))))))-.))).........((((.((((((((((......)))).)-))))).))))))).----------)-))))).. ( -30.10) >DroPer_CAF1 51590 92 - 1 CGUGUCACA----UCAUCGUUUUGUCCUUCUCUUUGACAAAACACAUG---------------UCAUCAUCUCGUGGGGCAUGCAAUGAUAAAGGAUGCUG-------CUGGCUGCCA ...((((((----.((((((((((((.........)))))))).((((---------------((.((((...))))))))))...........)))).))-------.))))..... ( -24.70) >consensus UGUGUCACAUAUAUCAUCGUUUUGUCUUCCUUUUUGACAAAGC_CAUGUCCACAUCACCGUCAUCAUCAUCCACACGAGGAUGU_AUGAUAAAGGAUGCUG_______CU_GAUGCCA .(((((............((((((((.........))))))))...................((((((((((......)))))..)))))....)))))................... (-17.29 = -17.68 + 0.39)

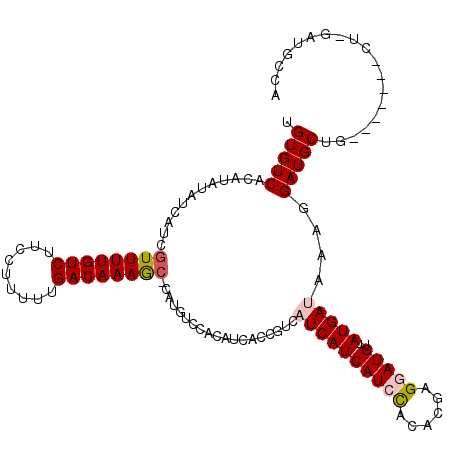

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:56 2006