
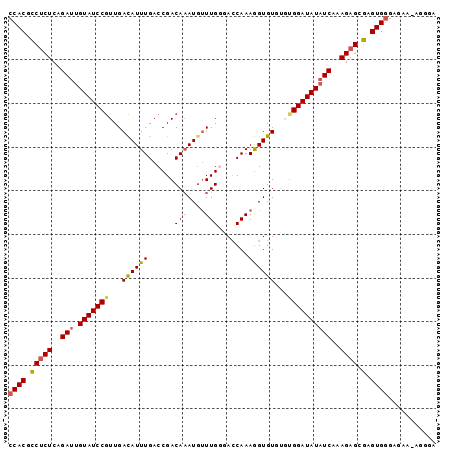
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,833,664 – 21,833,762 |
| Length | 98 |
| Max. P | 0.992381 |

| Location | 21,833,664 – 21,833,762 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21833664 98 + 23771897 CCACGCCCCUCAGAUUGUAUCCGUUGACAUUUGACCGACAAAUGUUUGGGACCAAAGGUGUGUGUGGAUAUAUCAAAGAGCGAGUGGGAGAAGAGGGA ((.(.((((((...(((..((((..((((((((.....))))))))))))..))).(((((((....))))))).......))).)))....).)).. ( -26.40) >DroSec_CAF1 14521 98 + 1 CCACGCCUCUCAGAUUGUAUCCGUUGACAUUUGACCGACAAAUGUUUGUUACCAAAGGUGUGUGUGGAUAUAUCAAAGAGCGAGUGGGAGAAAAGGGA ((((.(((((..(((.(((((((..((((((((.....))))))))...((((...))))....))))))))))..)))).).))))........... ( -27.80) >DroSim_CAF1 13195 97 + 1 CCACGCCUCUCAGAUUGUAUCCGUUGACAUUUGACCGACA-AUAUUUGUUACCAAAGGUGUGUGUGGAUAUAUCAAAGAGCGAGUGGGAGAAAAGGGA ((((.(((((..(((.(((((((...((((((....((((-.....)))).....))))))...))))))))))..)))).).))))........... ( -25.60) >DroEre_CAF1 13653 85 + 1 CCACGCCUCUCAGAUUGUAUCCGUUGACAUUUGACCGACAAAUGUUUGGGAGCAAAGGUGU----GGAUAUAUCAAAGAGAGAGUGGGA--------- ((((..(((((...((((.((((..((((((((.....)))))))))))).)))).(((((----....)))))...))))).))))..--------- ( -29.30) >DroYak_CAF1 8583 92 + 1 CCACAACUCUCAGAUUGUAUCCAUUGAUAUUUGACCGACAAAUGCUUGGAACCAAAAGUGU---UGGAUAUCUCAAAGAGAUAGUGAGAGAA---GGA ......((((((....(((((....)))))....((((((.....(((....)))...)))---))).((((((...)))))).))))))..---... ( -22.20) >consensus CCACGCCUCUCAGAUUGUAUCCGUUGACAUUUGACCGACAAAUGUUUGGGACCAAAGGUGUGUGUGGAUAUAUCAAAGAGCGAGUGGGAGAA_AGGGA ((((.(((((..(((.(((((((...((((((......(((....))).......))))))...))))))))))..)))).).))))........... (-20.68 = -20.92 + 0.24)


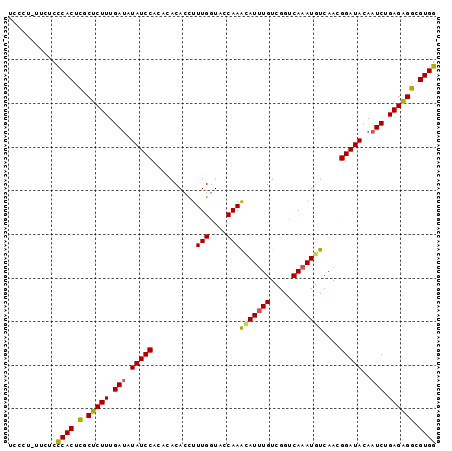
| Location | 21,833,664 – 21,833,762 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.14 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21833664 98 - 23771897 UCCCUCUUCUCCCACUCGCUCUUUGAUAUAUCCACACACACCUUUGGUCCCAAACAUUUGUCGGUCAAAUGUCAACGGAUACAAUCUGAGGGGCGUGG ...........((((..((((((((((.(((((..........(((....)))(((((((.....)))))))....)))))..))).))))))))))) ( -25.00) >DroSec_CAF1 14521 98 - 1 UCCCUUUUCUCCCACUCGCUCUUUGAUAUAUCCACACACACCUUUGGUAACAAACAUUUGUCGGUCAAAUGUCAACGGAUACAAUCUGAGAGGCGUGG ...........((((.(.(((((.(((.(((((......(((...))).....(((((((.....)))))))....)))))..))).)))))).)))) ( -25.00) >DroSim_CAF1 13195 97 - 1 UCCCUUUUCUCCCACUCGCUCUUUGAUAUAUCCACACACACCUUUGGUAACAAAUAU-UGUCGGUCAAAUGUCAACGGAUACAAUCUGAGAGGCGUGG ...........((((.(.(((((.(((.(((((.........((((....))))..(-((.((......)).))).)))))..))).)))))).)))) ( -21.40) >DroEre_CAF1 13653 85 - 1 ---------UCCCACUCUCUCUUUGAUAUAUCC----ACACCUUUGCUCCCAAACAUUUGUCGGUCAAAUGUCAACGGAUACAAUCUGAGAGGCGUGG ---------..((((((((((...(((.(((((----......(((....)))(((((((.....)))))))....)))))..))).)))))).)))) ( -24.50) >DroYak_CAF1 8583 92 - 1 UCC---UUCUCUCACUAUCUCUUUGAGAUAUCCA---ACACUUUUGGUUCCAAGCAUUUGUCGGUCAAAUAUCAAUGGAUACAAUCUGAGAGUUGUGG ...---......(((...(((((.((..((((((---.....((((....)))).(((((.....))))).....))))))...)).)))))..))). ( -20.10) >consensus UCCCU_UUCUCCCACUCGCUCUUUGAUAUAUCCACACACACCUUUGGUACCAAACAUUUGUCGGUCAAAUGUCAACGGAUACAAUCUGAGAGGCGUGG ...........((((.(.(((((.(((.(((((..........(((....)))(((((((.....)))))))....)))))..))).)))))).)))) (-21.46 = -21.14 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:53 2006