
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,821,946 – 21,822,082 |
| Length | 136 |
| Max. P | 0.923867 |

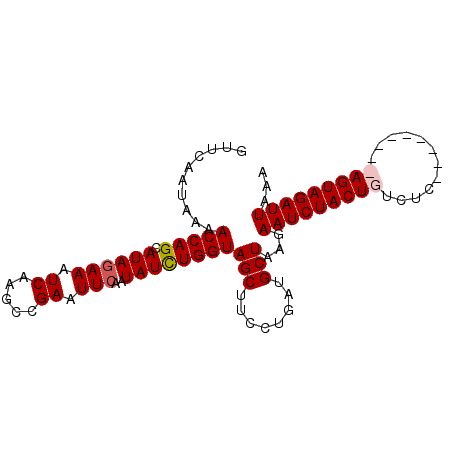
| Location | 21,821,946 – 21,822,042 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
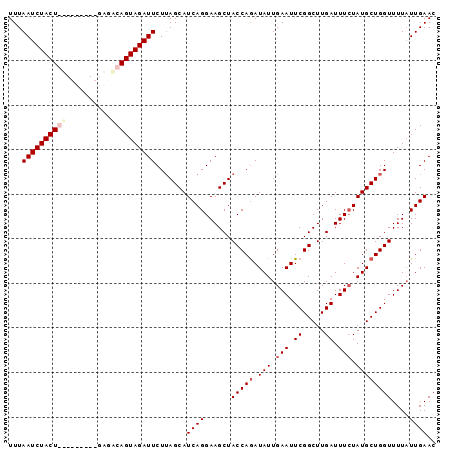
>3L_DroMel_CAF1 21821946 96 + 23771897 GUUCAAUAAAACCAGCAUAGAAAUCAAGCCGAAUUCAAUAUUUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUCACGCAAGACAGUAGAUUAAA .............(((((.........(((((((.....))))))).........)))))...((((((((((((......))))))))))))... ( -27.77) >DroSec_CAF1 1989 87 + 1 GUUCAAUAAAACCAGCAUAGAAAUCAAGCCGAAUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC---------AGUAGAUUAAA ..........(((((.((((((.((.....)).)))..))))))))(((........)))...(((((((((...)---------))))))))... ( -19.20) >DroSim_CAF1 2049 87 + 1 UUUCAAUAAAACCAGCAUAGAAAUCAAGCCGAAUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC---------AGUAGAUUAAA ..........(((((.((((((.((.....)).)))..))))))))(((........)))...(((((((((...)---------))))))))... ( -19.20) >DroEre_CAF1 1992 95 + 1 GUUCAA-AAAACCAGCAUACAAAUCAAGCCGAAUUCAAUAUCUGGUAGCUUCCUGAUGCUUAGAAUCUACUGCAUCACGCAUGACAGUAGAUUAAA ......-......(((((.........((((.((.....)).)))).........)))))...((((((((((((.....))).)))))))))... ( -19.07) >consensus GUUCAAUAAAACCAGCAUAGAAAUCAAGCCGAAUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC_________AGUAGAUUAAA ..........(((((.((((((.((.....)).)))..))))))))(((........)))...(((((((((............)))))))))... (-15.81 = -16.38 + 0.56)


| Location | 21,821,946 – 21,822,042 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -16.49 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21821946 96 - 23771897 UUUAAUCUACUGUCUUGCGUGAGACAGUAGAUUCUUAGCAUCAGGAAGCUACCAAAUAUUGAAUUCGGCUUGAUUUCUAUGCUGGUUUUAUUGAAC ...(((((((((((((....))))))))))))).(((((((.(((((....((.(((.....))).)).....))))))))))))........... ( -27.40) >DroSec_CAF1 1989 87 - 1 UUUAAUCUACU---------GAGACAGUAGAUUCUUAGCAUCAGGAAGCUACCAGAUAUUGAAUUCGGCUUGAUUUCUAUGCUGGUUUUAUUGAAC ...((((((((---------(...))))))))).......((((......(((((.(((.(((.((.....)).))).))))))))....)))).. ( -20.30) >DroSim_CAF1 2049 87 - 1 UUUAAUCUACU---------GAGACAGUAGAUUCUUAGCAUCAGGAAGCUACCAGAUAUUGAAUUCGGCUUGAUUUCUAUGCUGGUUUUAUUGAAA ...((((((((---------(...))))))))).......((((......(((((.(((.(((.((.....)).))).))))))))....)))).. ( -20.30) >DroEre_CAF1 1992 95 - 1 UUUAAUCUACUGUCAUGCGUGAUGCAGUAGAUUCUAAGCAUCAGGAAGCUACCAGAUAUUGAAUUCGGCUUGAUUUGUAUGCUGGUUUU-UUGAAC ...(((((((((.(((.....)))))))))))).......(((((((...(((((....((((((......))))))....))))))))-)))).. ( -23.50) >consensus UUUAAUCUACU_________GAGACAGUAGAUUCUUAGCAUCAGGAAGCUACCAGAUAUUGAAUUCGGCUUGAUUUCUAUGCUGGUUUUAUUGAAC ...((((((((((..........)))))))))).......((((......(((((.(((.(((.((.....)).))).))))))))....)))).. (-16.49 = -17.93 + 1.44)


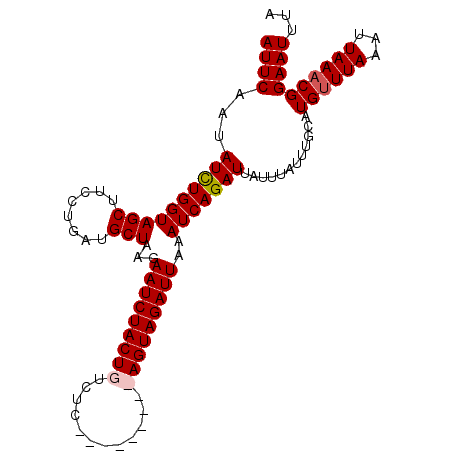
| Location | 21,821,978 – 21,822,082 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -16.46 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
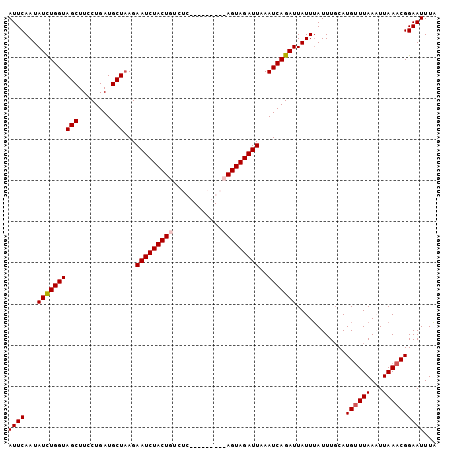
>3L_DroMel_CAF1 21821978 104 + 23771897 AUUCAAUAUUUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUCACGCAAGACAGUAGAUUAAAUCAGAUUAUUUAUUUGCAUGUUUAAAUUAAACGGAAUUUA ((((...((((((((((........)))...((((((((((((......))))))))))))..)))))))............((((((...))))))))))... ( -24.50) >DroSec_CAF1 2021 95 + 1 AUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC---------AGUAGAUUAAAUCAGAUUAUUUAUUUGCAUGUUUAACUUAAACGGAAUUUA ((((...((((((((((........)))...(((((((((...)---------))))))))..)))))))............((((((...))))))))))... ( -19.90) >DroSim_CAF1 2081 95 + 1 AUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC---------AGUAGAUUAAAUCAGAUUAUUUAUUUGCAUGUUUAACUUAAACGGAAUUUA ((((...((((((((((........)))...(((((((((...)---------))))))))..)))))))............((((((...))))))))))... ( -19.90) >DroEre_CAF1 2023 104 + 1 AUUCAAUAUCUGGUAGCUUCCUGAUGCUUAGAAUCUACUGCAUCACGCAUGACAGUAGAUUAAAUCAGAUUAUUAAUUUCCAUGCUUAAAUUAAUCGGAAUUUU ((((...((((((((((........)))...((((((((((((.....))).)))))))))..))))))).((((((((........))))))))..))))... ( -22.50) >consensus AUUCAAUAUCUGGUAGCUUCCUGAUGCUAAGAAUCUACUGUCUC_________AGUAGAUUAAAUCAGAUUAUUUAUUUGCAUGUUUAAAUUAAACGGAAUUUA ((((...((((((((((........)))...(((((((((............)))))))))..)))))))............((((((...))))))))))... (-16.46 = -17.02 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:48 2006