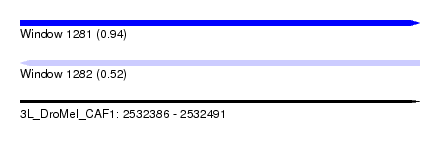
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,532,386 – 2,532,491 |
| Length | 105 |
| Max. P | 0.940319 |

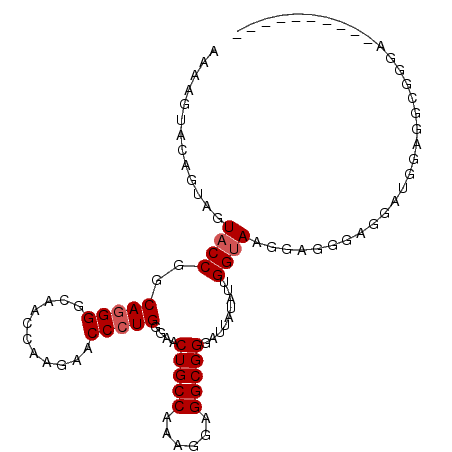
| Location | 2,532,386 – 2,532,491 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
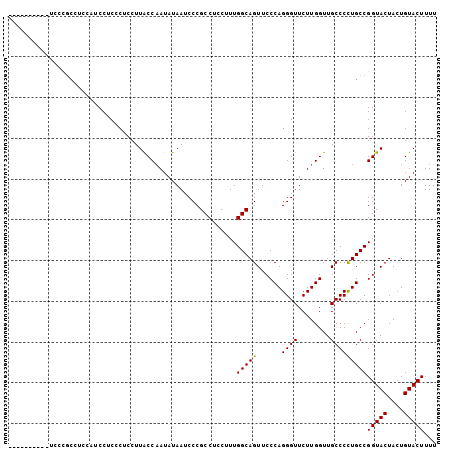
>3L_DroMel_CAF1 2532386 105 + 23771897 UGUCCCUCUGUCCUGCCUCCAUCCUCCCUCCUUACCAAUAUAAUCCCGCCUCCUUUGGCAGUUCCCAGGGUUCUUGGUUGCCCCUGCCGGUACUACUGUACUUUU ...((((..(..(((((.......................................)))))..)..)))).....(((.......)))(((((....)))))... ( -19.43) >DroSec_CAF1 155256 98 + 1 -------CUGUCCCGCCUCCAACCUCCCUCCUUACCAAUAUAAUCCCGCCUCCUUUGGCAGUUCCCAGGGUUCUUGGUUGCCCCUGCCGGUACUACUGUACUUUU -------.......((..((((...((((..................(((......))).(....)))))...))))..)).......(((((....)))))... ( -19.30) >DroSim_CAF1 152819 95 + 1 ----------UUCCGCCUCCAUCCUCCCUCCUUGCCAAUAUAAUCCCGCCUCCUUUGGCAGUUCCCAGGGUUCUUGGUUGCCCCUGCCGGUACUACUGUACUUUU ----------....((..(((....((((..(((((((................))))))).....))))....)))..)).......(((((....)))))... ( -22.29) >DroYak_CAF1 155471 83 + 1 ----------------------CCUCACUCCUUACCUAUAUAAUCCCGCCUCCUUUGGCAGUUCCCAGGGUUCUUGGUUGCCCUUGCCGGUACUACUGUACUUUU ----------------------..................................((((((..(((((...)))))..))...))))(((((....)))))... ( -15.80) >consensus __________UCCCGCCUCCAUCCUCCCUCCUUACCAAUAUAAUCCCGCCUCCUUUGGCAGUUCCCAGGGUUCUUGGUUGCCCCUGCCGGUACUACUGUACUUUU ........................................................(((((......((((........)))))))))(((((....)))))... (-16.29 = -16.10 + -0.19)

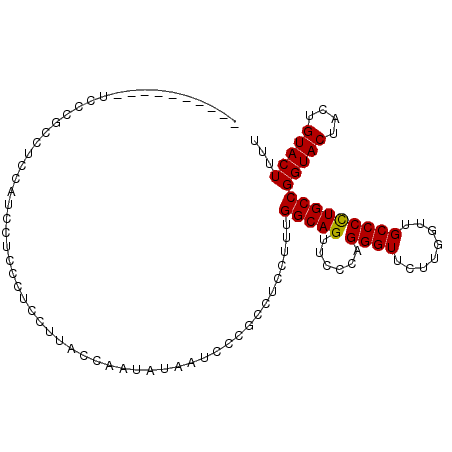
| Location | 2,532,386 – 2,532,491 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2532386 105 - 23771897 AAAAGUACAGUAGUACCGGCAGGGGCAACCAAGAACCCUGGGAACUGCCAAAGGAGGCGGGAUUAUAUUGGUAAGGAGGGAGGAUGGAGGCAGGACAGAGGGACA ....((.(..(.((.((.((..((....)).....((((.....(((((......)))))..((((....))))..)))).........)).)))).)..).)). ( -24.20) >DroSec_CAF1 155256 98 - 1 AAAAGUACAGUAGUACCGGCAGGGGCAACCAAGAACCCUGGGAACUGCCAAAGGAGGCGGGAUUAUAUUGGUAAGGAGGGAGGUUGGAGGCGGGACAG------- ....((...((....(((((..((....)).....((((.....(((((......)))))..((((....))))..))))..)))))..))...))..------- ( -26.20) >DroSim_CAF1 152819 95 - 1 AAAAGUACAGUAGUACCGGCAGGGGCAACCAAGAACCCUGGGAACUGCCAAAGGAGGCGGGAUUAUAUUGGCAAGGAGGGAGGAUGGAGGCGGAA---------- ...............(((.(..((....)).....(((((....)((((((..((.......))...))))))...))))........).)))..---------- ( -22.90) >DroYak_CAF1 155471 83 - 1 AAAAGUACAGUAGUACCGGCAAGGGCAACCAAGAACCCUGGGAACUGCCAAAGGAGGCGGGAUUAUAUAGGUAAGGAGUGAGG---------------------- .......((.(..((((.....((....)).....(((.(....).(((......))))))........))))...).))...---------------------- ( -20.30) >consensus AAAAGUACAGUAGUACCGGCAGGGGCAACCAAGAACCCUGGGAACUGCCAAAGGAGGCGGGAUUAUAUUGGUAAGGAGGGAGGAUGGAGGCGGGA__________ .............((((..(((((...........)))))....(((((......))))).........))))................................ (-17.92 = -18.43 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:19 2006