
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,810,257 – 21,810,395 |
| Length | 138 |
| Max. P | 0.982160 |

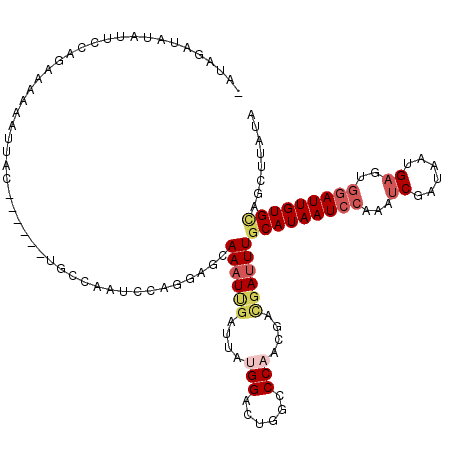
| Location | 21,810,257 – 21,810,369 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.97 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21810257 112 + 23771897 -ACAGAUAUAUUCCAGCAAAA-UUUC------UGCCAAUCCAGAAGUAAAUUGAUUAUGGACUGGCCCAACGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCGGAUUGUG -.(((......((((.(((.(-((((------((......)))))))...)))....)))))))........(((((((((((((((((..((......)).))))))))))))))))). ( -33.20) >DroSec_CAF1 30352 114 + 1 AAUGGAUAUAUUCCAGCAAAAGUUAC------UGCCAAUCCAGGAGCAAAUUGAUUAUGGACUGGCCCAGCGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGGAGCUUAUA .....(((..((((.(((((.((..(------(((((.((((....(.....)....)))).))))..))..))..)))))((((((((..((......)).))))))))))))..))). ( -32.20) >DroSim_CAF1 35803 114 + 1 AAUGGAUAUAUUCCAGCAAAAGUUAC------UGCCAAUCCAGGAGCAAAUUGAUUAUGGACUGGCCCAGCGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUA ..((((.....))))((....((..(------(((((.((((....(.....)....)))).))))..))..))....(((((((((((..((......)).)))))))))))))..... ( -35.30) >DroEre_CAF1 37617 113 + 1 -AUAGAUAGAUUCGAGAAAAAAUUCC------CGUCAAUCCAGGAGCAAAUCGAUUUGGGACUCGCCCAACGACGAUUUGCAUAAUCAAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUA -...(((.((..((.(((....))).------)))).)))...((((((((((..(((((.....)))))...))))))((((((((....((......))...)))))))).))))... ( -28.70) >DroYak_CAF1 46206 104 + 1 GAUAGAUAGAUUCAAGAAGAAA----------------UCCAGGACCAAAUUGAUAAGGGAGUCACCCAGCGAUGAUUUGCAUAAUCCAAAACGAUAAUGAGUGGAUUGUGCAGCUUAUA ........(((((..(.(....----------------).)....((..........)))))))........((((.((((((((((((...(......)..)))))))))))).)))). ( -20.70) >DroAna_CAF1 44917 115 + 1 -GUAGUU---UUCGAAAAAAUCGUCCGACAUUAGGCAAUAAAAAUUCAACUGGAUUAUGGAGC-GUCCAAUGCGGAUUUGCAUAAUCCGAAUCGAUAAUGAGGGAAUUGUGUGGUGGCUA -.((((.---..(((.....))).....(((((.(((((...........(((((((((.((.-((((.....)))))).)))))))))..((......))....))))).))))))))) ( -24.90) >consensus _AUAGAUAUAUUCCAGAAAAAAUUAC______UGCCAAUCCAGGAGCAAAUUGAUUAUGGACUGGCCCAACGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUA ...............................................((((((....(((......)))....))))))(((((((((...((......))..)))))))))........ (-12.80 = -13.97 + 1.17)

| Location | 21,810,257 – 21,810,369 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -10.10 |
| Energy contribution | -11.47 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21810257 112 - 23771897 CACAAUCCGCACAAUCCACUCAUUAUCGAUUUGGAUUAUGCAAAUCGUCGUUGGGCCAGUCCAUAAUCAAUUUACUUCUGGAUUGGCA------GAAA-UUUUGCUGGAAUAUAUCUGU- .....(((....((((((.((......))..))))))..(((((...((.....(((((((((...............))))))))).------))..-.))))).)))..........- ( -27.96) >DroSec_CAF1 30352 114 - 1 UAUAAGCUCCACAAUCCACUCAUUAUCGAUUUGGAUUAUGCAAAUCGUCGCUGGGCCAGUCCAUAAUCAAUUUGCUCCUGGAUUGGCA------GUAACUUUUGCUGGAAUAUAUCCAUU .......((((.((((((.((......))..))))))..(((((..((..((..(((((((((...............))))))))))------)..)).)))))))))........... ( -29.26) >DroSim_CAF1 35803 114 - 1 UAUAAGCUGCACAAUCCACUCAUUAUCGAUUUGGAUUAUGCAAAUCGUCGCUGGGCCAGUCCAUAAUCAAUUUGCUCCUGGAUUGGCA------GUAACUUUUGCUGGAAUAUAUCCAUU ....(((((((.((((((.((......))..)))))).))))....((..((..(((((((((...............))))))))))------)..))....)))(((.....)))... ( -31.56) >DroEre_CAF1 37617 113 - 1 UAUAAGCUGCACAAUCCACUCAUUAUCGAUUUUGAUUAUGCAAAUCGUCGUUGGGCGAGUCCCAAAUCGAUUUGCUCCUGGAUUGACG------GGAAUUUUUUCUCGAAUCUAUCUAU- ...........(((((((......((((....))))...((((((((...(((((.....)))))..))))))))...))))))).((------((((....))))))...........- ( -32.50) >DroYak_CAF1 46206 104 - 1 UAUAAGCUGCACAAUCCACUCAUUAUCGUUUUGGAUUAUGCAAAUCAUCGCUGGGUGACUCCCUUAUCAAUUUGGUCCUGGA----------------UUUCUUCUUGAAUCUAUCUAUC .(((((.((((.((((((..(......)...)))))).))))..(((((....)))))....))))).....(((...((((----------------(((......))))))).))).. ( -19.00) >DroAna_CAF1 44917 115 - 1 UAGCCACCACACAAUUCCCUCAUUAUCGAUUCGGAUUAUGCAAAUCCGCAUUGGAC-GCUCCAUAAUCCAGUUGAAUUUUUAUUGCCUAAUGUCGGACGAUUUUUUCGAA---AACUAC- ...............(((..(((((((((((.((((((((....(((.....))).-....))))))))))))))............)))))..)))(((.....)))..---......- ( -22.40) >consensus UAUAAGCUGCACAAUCCACUCAUUAUCGAUUUGGAUUAUGCAAAUCGUCGCUGGGCCAGUCCAUAAUCAAUUUGCUCCUGGAUUGGCA______GGAACUUUUGCUGGAAUAUAUCUAU_ ...........(((((((.........((((((.......))))))......(((....)))................)))))))................................... (-10.10 = -11.47 + 1.36)

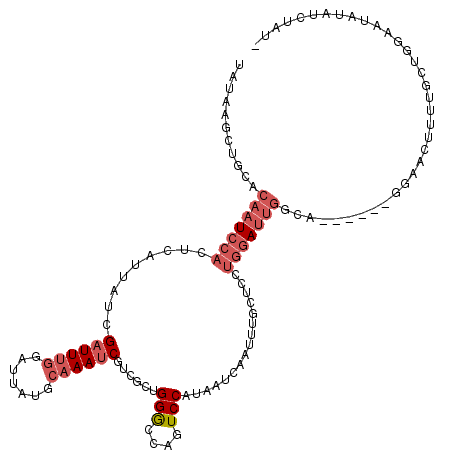

| Location | 21,810,289 – 21,810,395 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -22.24 |
| Energy contribution | -23.72 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21810289 106 + 23771897 CAGAAGUAAAUUGAUUAUGGACUGGCCCAACGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCGGAUUGUGAGUGGGCGGA----------GUG-GCAUGCCACUGAA--- (((..((((.....))))...)))(((((...(((((((((((((((((..((......)).)))))))))))))))))...)))))..(----------(((-(....)))))...--- ( -42.30) >DroSec_CAF1 30386 107 + 1 CAGGAGCAAAUUGAUUAUGGACUGGCCCAGCGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGGAGCUUAUAAGUGGGCGGA----------GUGGGCAUACCACUGAA--- .....((((((((....(((......)))....))))))))((((((((..((......)).))))))))...(((((....)))))..(----------((((.....)))))...--- ( -31.20) >DroSim_CAF1 35837 107 + 1 CAGGAGCAAAUUGAUUAUGGACUGGCCCAGCGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUAAGUGGGCGGA----------GUGGGCAUACCACUGAA--- (((..................)))(((((...(..(.((((((((((((..((......)).)))))))))))).)..)...)))))..(----------((((.....)))))...--- ( -33.27) >DroEre_CAF1 37650 116 + 1 CAGGAGCAAAUCGAUUUGGGACUCGCCCAACGACGAUUUGCAUAAUCAAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUAAGUGGGCGGAG-UGGGCGGAGUGGGCGGACCACUGAA--- (((((.....))....(((...(((((((.((.(.((((((((((((....((......))...)))))))).(((((....))))).)))-).).))...))))))).))))))..--- ( -37.30) >DroYak_CAF1 46230 107 + 1 CAGGACCAAAUUGAUAAGGGAGUCACCCAGCGAUGAUUUGCAUAAUCCAAAACGAUAAUGAGUGGAUUGUGCAGCUUAUAAGUGGGCGGA----------GUGGUCCGACCACUGAA--- ..((((((...((((......))))((((...((((.((((((((((((...(......)..)))))))))))).))))...))))....----------.))))))..........--- ( -35.50) >DroAna_CAF1 44953 119 + 1 AAAAUUCAACUGGAUUAUGGAGC-GUCCAAUGCGGAUUUGCAUAAUCCGAAUCGAUAAUGAGGGAAUUGUGUGGUGGCUAAGUGGGUUGCGAUUUGCGGGGGAGGCAGGUGACAGAUCCC ...........(((((.....((-.(((..((((((((.(((((((((...((......)).)).))))))).(..(((.....)))..)))))))))..))).)).(....).))))). ( -36.70) >consensus CAGGAGCAAAUUGAUUAUGGACUGGCCCAACGACGAUUUGCAUAAUCCAAAUCGAUAAUGAGUGGAUUGUGCAGCUUAUAAGUGGGCGGA__________GUGGGCAGACCACUGAA___ ........................(((((...((((.(((((((((((...((......))..))))))))))).))))...))))).............((((.....))))....... (-22.24 = -23.72 + 1.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:43 2006