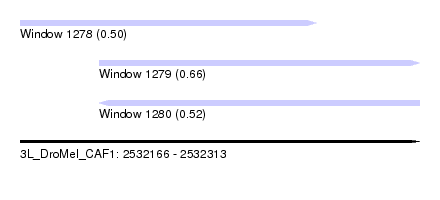
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,532,166 – 2,532,313 |
| Length | 147 |
| Max. P | 0.664879 |

| Location | 2,532,166 – 2,532,275 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -24.20 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
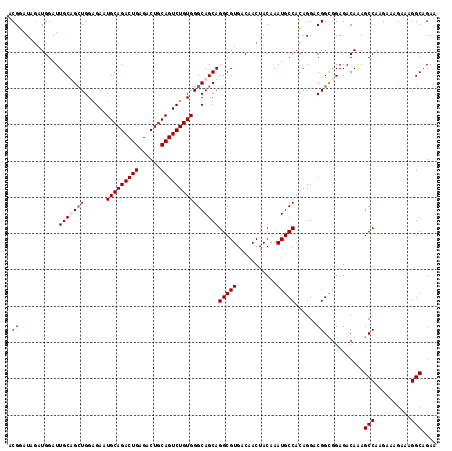
>3L_DroMel_CAF1 2532166 109 + 23771897 U-G----------GAAAUAUAUACAAAUUUCUCCCCCUGUACGGAUAGAUGGAUUGCUGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCA .-.----------(((((........)))))(((..((((....))))..))).(((((((......(((((((((......))))))))))))))))(((((...........))))). ( -33.30) >DroSec_CAF1 155026 120 + 1 CUGGGAAUUUAAAAAGAUAUAUAGAAAUUUCUCCCCCUGUACGGAUAGAUGGAUUGCUGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCA ...(((((((..............)))))))(((..((((....))))..))).(((((((......(((((((((......))))))))))))))))(((((...........))))). ( -33.44) >DroSim_CAF1 152594 120 + 1 AUGCGAAUUUAAAAAGAUAUAUAGAAAUUUCUCCCCCUGUACGGAUAGAUGGAUUGCAGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCA .(((........................((((((..(((((.............)))))..)))))).((((((((......)))))))).....)))(((((...........))))). ( -32.72) >DroYak_CAF1 155242 120 + 1 AUGGAAAUUUGUCCAGAUAUAUAUAAAUUUCGCGCACUGUACGUAUAUAUGGAUUGCAGAUGGGGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCA .((((......))))..............((((((.((((..((...((((((((((((.(.((.........)).)..)))))))))))).)).))))))))))............... ( -34.40) >consensus AUGGGAAUUUAAAAAGAUAUAUAGAAAUUUCUCCCCCUGUACGGAUAGAUGGAUUGCAGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCA ...............................(((..((((....))))..))).(((((((......(((((((((......))))))))))))))))(((((...........))))). (-24.20 = -25.70 + 1.50)

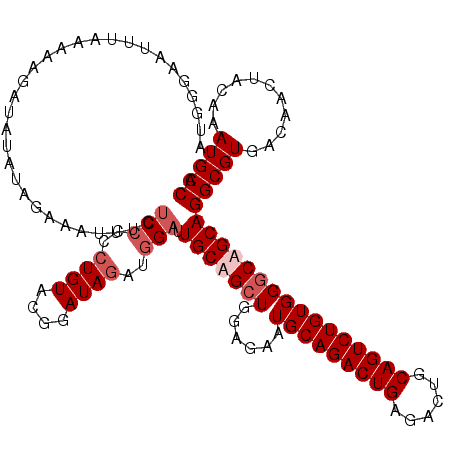
| Location | 2,532,195 – 2,532,313 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -28.08 |
| Energy contribution | -28.83 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
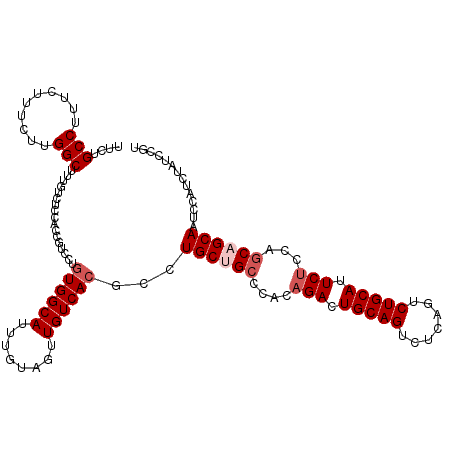
>3L_DroMel_CAF1 2532195 118 + 23771897 ACGGAUAGAUGGAUUGCUGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCACAGGACGGUGGAGACAGAGCCAAGAAAGAAAGGCUGAA .((...........(((((((......(((((((((......))))))))))))))))(((((...........)))))......)).((....)).((((..........))))... ( -35.80) >DroSec_CAF1 155066 118 + 1 ACGGAUAGAUGGAUUGCUGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCACAGGACGGCGGAGACAAAGCCAAGAAAGAAAGGCAGAA .((...........(((((((......(((((((((......))))))))))))))))(((((...........)))))......))..(....)...(((..........))).... ( -34.40) >DroSim_CAF1 152634 118 + 1 ACGGAUAGAUGGAUUGCAGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCACAGGACGGCCGAGACAAAGCCAAGAAAGAAAGGCAGAA .(((.....((..(((..((((.....(((((((((......)))))))))..)))).(((((...........))))).)))..)).))).......(((..........))).... ( -33.40) >DroYak_CAF1 155282 118 + 1 ACGUAUAUAUGGAUUGCAGAUGGGGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCAAAGGACGGUGGAGACAAAGCCAAGCGAGAAAGGCAGAA ..((...((((((((((((.(.((.........)).)..)))))))))))).)).((.(((((...........)))))...((.(..((....))..)))..))............. ( -33.30) >consensus ACGGAUAGAUGGAUUGCAGCUGGAGAAUGCAGACUGAGACUGCAGUCUGUGGGCAGCAGGCGUGACAACUACAAAUGCCACAGGACGGCGGAGACAAAGCCAAGAAAGAAAGGCAGAA .((...........(((((((......(((((((((......))))))))))))))))(((((...........)))))......))...........(((..........))).... (-28.08 = -28.83 + 0.75)

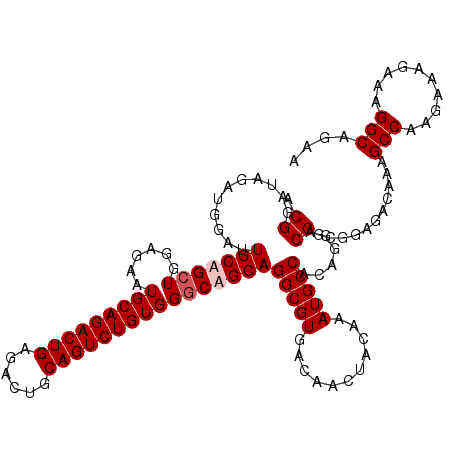
| Location | 2,532,195 – 2,532,313 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -23.15 |
| Energy contribution | -24.40 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
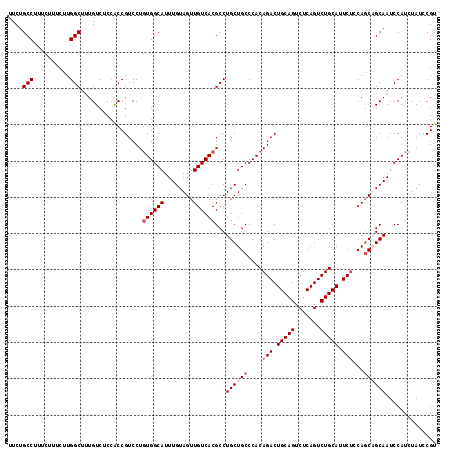
>3L_DroMel_CAF1 2532195 118 - 23771897 UUCAGCCUUUCUUUCUUGGCUCUGUCUCCACCGUCCUGUGGCAUUUGUAGUUGUCACGCCUGCUGCCCACAGACUGCAGUCUCAGUCUGCAUUCUCCAGCAGCAAUCCAUCUAUCCGU ...((((..........))))................((((((........))))))...((((((....(((.(((((.......))))).)))...)))))).............. ( -30.00) >DroSec_CAF1 155066 118 - 1 UUCUGCCUUUCUUUCUUGGCUUUGUCUCCGCCGUCCUGUGGCAUUUGUAGUUGUCACGCCUGCUGCCCACAGACUGCAGUCUCAGUCUGCAUUCUCCAGCAGCAAUCCAUCUAUCCGU ....((...........(((.........))).....((((((........)))))))).((((((....(((.(((((.......))))).)))...)))))).............. ( -29.90) >DroSim_CAF1 152634 118 - 1 UUCUGCCUUUCUUUCUUGGCUUUGUCUCGGCCGUCCUGUGGCAUUUGUAGUUGUCACGCCUGCUGCCCACAGACUGCAGUCUCAGUCUGCAUUCUCCAGCUGCAAUCCAUCUAUCCGU ...((((...(.....(((((.......)))))....).)))).(((((((((....((.....))...(((((((......))))))).......)))))))))............. ( -29.00) >DroYak_CAF1 155282 118 - 1 UUCUGCCUUUCUCGCUUGGCUUUGUCUCCACCGUCCUUUGGCAUUUGUAGUUGUCACGCCUGCUGCCCACAGACUGCAGUCUCAGUCUGCAUUCCCCAUCUGCAAUCCAUAUAUACGU ....(((..........)))...........(((....(((...((((((.((....((.....))...(((((((......))))))).......)).)))))).))).....))). ( -22.70) >consensus UUCUGCCUUUCUUUCUUGGCUUUGUCUCCACCGUCCUGUGGCAUUUGUAGUUGUCACGCCUGCUGCCCACAGACUGCAGUCUCAGUCUGCAUUCUCCAGCAGCAAUCCAUCUAUCCGU ....(((..........))).................((((((........))))))...((((((....(((.(((((.......))))).)))...)))))).............. (-23.15 = -24.40 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:17 2006