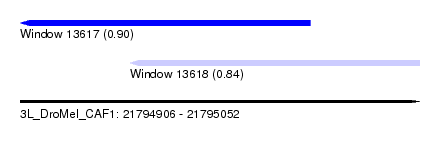
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,794,906 – 21,795,052 |
| Length | 146 |
| Max. P | 0.901907 |

| Location | 21,794,906 – 21,795,012 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -9.23 |
| Energy contribution | -11.68 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21794906 106 - 23771897 GGUUGGCCACCGGAGAUUGGUAUGCAAUUUCUUGGCCAACAAAAGCCCAA--CUUUA------CUAGGAAGGGUAAAGAGUAAUGAAUAUAUUUCGAACAACCAAAUUAUGCAA .((((((((..((((((((.....))))))))))))))))....((....--(((((------((......))))))).....((((.....))))..............)).. ( -30.10) >DroSec_CAF1 14748 106 - 1 GGUUGGCCACCGGAGAUUGGUAUGCAAUUUCUGGGCCAACAAAAGUCCAA--CUUUA------CAAGGAAGGGUAAAGAGUAAUGAAUAUAUUUCGAACCUUCAAAUCAUGCUA .(((((((..(((((((((.....)))))))))))))))).((((.....--)))).------....(((((.....(((((.......)))))....)))))........... ( -29.40) >DroSim_CAF1 20170 106 - 1 GGUUGGCCACCGGAGAUUGGUAUGCAAUUUCUUGGCCAACAAAAGCCCAA--CUUUA------CAAGGAAGGGUAAAGAGUAAUGAAUAUAUUUCGAACCAUCAAAUCAUGCUA .((((((((..((((((((.....))))))))))))))))....((((..--(((..------..)))..))))....((((..(((.....)))((....))......)))). ( -29.80) >DroEre_CAF1 21521 95 - 1 ------G-ACAGGCAAAGGGUAUGCAAUUUCUUAGCCA-UCAAAGCCCAA--CUUUA------CAAGUAAGUGGUAAAAGUAACGAAUAUACUUCGAACCACAAAAUUAUU--- ------.-...(((.(((((........))))).))).-...........--.....------.......(((((..(((((.......)))))...))))).........--- ( -16.90) >DroYak_CAF1 29605 107 - 1 ------CCACCGGAGAUUUGU-UAAAAUUUCUUAGCCAACCCAAGCCCAAGCCCUUAGAAGCCCAAGCAAGGCUAAAAAGUAAUUAAUAUAUUUCAAACCUCCAAAUUAUGCUA ------......(((((.(((-(((.((((.((((((.......((....))........((....))..)))))).))))..)))))).)))))................... ( -17.20) >consensus GGUUGGCCACCGGAGAUUGGUAUGCAAUUUCUUGGCCAACAAAAGCCCAA__CUUUA______CAAGGAAGGGUAAAGAGUAAUGAAUAUAUUUCGAACCACCAAAUUAUGCUA .(((((((...((((((((.....)))))))).)))))))...........................................((((.....)))).................. ( -9.23 = -11.68 + 2.45)

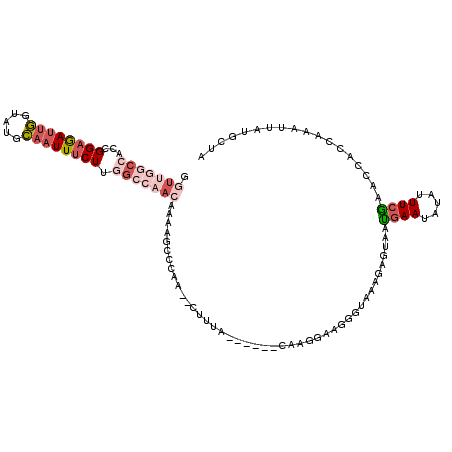
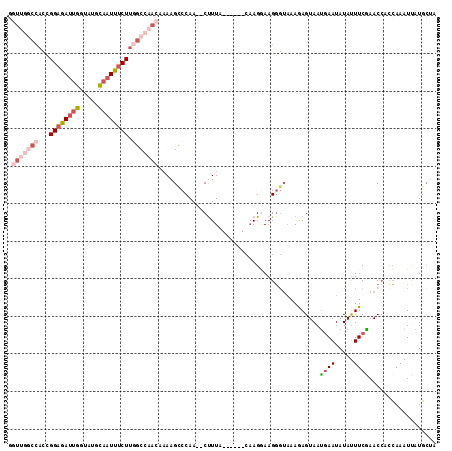
| Location | 21,794,946 – 21,795,052 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.38 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -6.53 |
| Energy contribution | -7.53 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21794946 106 - 23771897 AUAUUAAGGGAUAAUUUGUCACGAAAGGGACAGGCAAAGGGGUUGGCCACCGGAG-AUUGGUAUGCAAUUUCUUGGCCAA-CAAAAGC--CCAA----CUUUA------CUAGGAAGGGU .......(((....((((((.(....).)))))).......((((((((..((((-((((.....)))))))))))))))-).....)--))..----((((.------....))))... ( -35.10) >DroSec_CAF1 14788 106 - 1 AUAUUAAGGGGUAAUUUGUCACGAAAGGGACAGGCAAAGGGGUUGGCCACCGGAG-AUUGGUAUGCAAUUUCUGGGCCAA-CAAAAGU--CCAA----CUUUA------CAAGGAAGGGU .......(((.(..((((((.(....).)))))).......(((((((..(((((-((((.....)))))))))))))))-)...).)--))..----((((.------....))))... ( -33.20) >DroSim_CAF1 20210 106 - 1 AUAUUAAGGGGUAAUUUGUCACGAAAGGGACAGGCAAAGGGGUUGGCCACCGGAG-AUUGGUAUGCAAUUUCUUGGCCAA-CAAAAGC--CCAA----CUUUA------CAAGGAAGGGU ........((((..((((((.(....).)))))).......((((((((..((((-((((.....)))))))))))))))-)....))--))..----((((.------....))))... ( -36.60) >DroEre_CAF1 21558 85 - 1 AUAUUGAGGAGUAAUUCGUCACGAAAG-------------------G-ACAGGCA-AAGGGUAUGCAAUUUCUUAGCCA--UCAAAGC--CCAA----CUUUA------CAAGUAAGUGG ...(((((((....)))(((.(....)-------------------)-)).(((.-(((((........))))).))).--))))...--(((.----(((..------.....)))))) ( -19.00) >DroYak_CAF1 29645 94 - 1 AUAUUAAGGGGUAAUUUGUCACAAAAG-------------------CCACCGGAG-AUUUGU-UAAAAUUUCUUAGCCAA-CCCAAGC--CCAAGC--CCUUAGAAGCCCAAGCAAGGCU ...(((((((.(...............-------------------.....((((-((((..-..))))))))..((...-.....))--...).)--)))))).((((.......)))) ( -17.50) >DroAna_CAF1 25008 110 - 1 AUAUUAAGGGGUAAUUUGUCGCCGCAGGGACGGGGAGAAGGGGUAACCCUUACAGAAUUGGGAUGCAAUUUCUUAGCCCGACCAAGGUGGCCAAGAAAAAUUA------CAGGCAA---- ..........(((((((.(((((((..((.((((((((((..(((.(((..........))).)))..))))))..)))).))...)))))...)).))))))------)......---- ( -37.20) >consensus AUAUUAAGGGGUAAUUUGUCACGAAAGGGACAGGCAAAGGGGUUGGCCACCGGAG_AUUGGUAUGCAAUUUCUUAGCCAA_CAAAAGC__CCAA____CUUUA______CAAGGAAGGGU ...(((((((((...(((((.(....).)))))...............((((......)))).....)))))))))............................................ ( -6.53 = -7.53 + 1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:32 2006