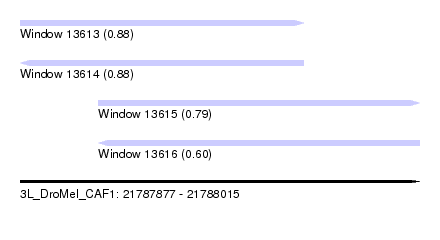
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,787,877 – 21,788,015 |
| Length | 138 |
| Max. P | 0.883462 |

| Location | 21,787,877 – 21,787,975 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21787877 98 + 23771897 --CACAAA---AUUUCGCUAGUGGGC----AAGCAA-----AAUAACGAGUAUUGGUUAAAAA-A-------AAAGUGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCA --(((...---.....(((.(....)----.)))..-----..((((........))))....-.-------...)))..(((((..(((((.......)))))..)))))......... ( -23.90) >DroSec_CAF1 7378 99 + 1 --CACAAU---AUUUCGCUGGUGGGC----AAAGAA-----AAUAACGAGUAUUGCUCAAAAAAA-------AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCA --......---.....((.(((((((----((....-----...........)))))).......-------........(((((..(((((.......)))))..)))))..))).)). ( -26.16) >DroSim_CAF1 12644 97 + 1 --CACAAU---AUUUCGCUGGUGGGC----AAACAA-----AAUAACGAGUAUUGCUCAAAAA---------AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCA --......---.....((.(((((((----((((..-----........)).)))))).....---------........(((((..(((((.......)))))..)))))..))).)). ( -27.10) >DroEre_CAF1 13478 92 + 1 --UACAGA---AUU-CGCUGGUGGGC----AAGCAA-----AAUAACGAGUAUCCCCUAAAAA-------------AGAAAGGGGCACCCGGAUUCGAACCGGGGACCUCUCGAUCUGUG --((((((---...-.(((.......----.)))..-----.....((((...(((((.....-------------....)))))..(((((.......))))).....)))).)))))) ( -29.10) >DroYak_CAF1 13381 90 + 1 --CACAAA---AUUUCGC---UGGGC----AAACAA-----AAAAACGAGUAUCCCUUAAAAA-------------AGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCA --......---.....((---((...----...)).-----.....((((...(((((.....-------------..)))))((..(((((.......)))))..)).))))....)). ( -24.60) >DroAna_CAF1 17840 120 + 1 CUGGCAGUAUAUUUUCGUUUAUGAGCGGGAAAGUAAUGGUUUAUAAAGAAUACCAUUAAAGGAAAAAAAGCCAAAAAAAAGGGGGCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCA .((((......((..((((....))))..))..(((((((...........)))))))...........)))).......(((((..((.((.......)).))..)))))......... ( -28.60) >consensus __CACAAA___AUUUCGCUGGUGGGC____AAACAA_____AAUAACGAGUAUCGCUUAAAAA_________AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCA ................(((....)))......................................................(((((..(((((.......)))))..)))))......... (-16.37 = -16.32 + -0.05)



| Location | 21,787,877 – 21,787,975 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -17.92 |
| Energy contribution | -16.82 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21787877 98 - 23771897 UGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCACUUU-------U-UUUUUAACCAAUACUCGUUAUU-----UUGCUU----GCCCACUAGCGAAAU---UUUGUG-- .(((((.((((.((..(((((.......)))))..)).........((-------.-.....))......))))..(((-----(((((.----.......))))))))---))))).-- ( -23.90) >DroSec_CAF1 7378 99 - 1 UGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU-------UUUUUUUGAGCAAUACUCGUUAUU-----UUCUUU----GCCCACCAGCGAAAU---AUUGUG-- .((((...((((((..(((((.......)))))..))...........-------.......(((.....))).....)-----)))(((----((......)))))..---.)))).-- ( -22.10) >DroSim_CAF1 12644 97 - 1 UGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU---------UUUUUGAGCAAUACUCGUUAUU-----UUGUUU----GCCCACCAGCGAAAU---AUUGUG-- .((((..((((.((..(((((.......)))))..)).....(((...---------.....))).....)))).((((-----(((((.----.......))))))))---))))).-- ( -24.20) >DroEre_CAF1 13478 92 - 1 CACAGAUCGAGAGGUCCCCGGUUCGAAUCCGGGUGCCCCUUUCU-------------UUUUUAGGGGAUACUCGUUAUU-----UUGCUU----GCCCACCAGCG-AAU---UCUGUA-- .(((((.((((.((..(((((.......)))))..))((((...-------------.....))))....))))....(-----(((((.----.......))))-)).---))))).-- ( -30.40) >DroYak_CAF1 13381 90 - 1 UGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCU-------------UUUUUAAGGGAUACUCGUUUUU-----UUGUUU----GCCCA---GCGAAAU---UUUGUG-- .(((((.((((.((..(((((.......)))))..))(((((..-------------.....)))))...))))...((-----(((((.----....)---)))))).---))))).-- ( -28.80) >DroAna_CAF1 17840 120 - 1 UGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGCCCCCUUUUUUUUGGCUUUUUUUCCUUUAAUGGUAUUCUUUAUAAACCAUUACUUUCCCGCUCAUAAACGAAAAUAUACUGCCAG .((((...((((((..(((((.......)))))..)).((........)).....))))...(((((((...........)))))))..........................))))... ( -24.00) >consensus UGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU_________UUUUUAAGCAAUACUCGUUAUU_____UUGCUU____GCCCACCAGCGAAAU___AUUGUG__ .((((..((((.((..(((((.......)))))..)).....................((......))..))))....................((......)).........))))... (-17.92 = -16.82 + -1.11)

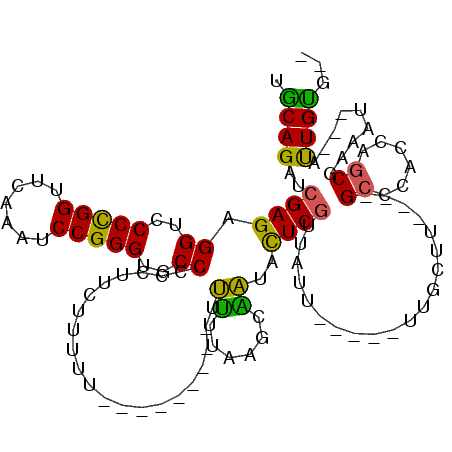
| Location | 21,787,904 – 21,788,015 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21787904 111 + 23771897 -AAUAACGAGUAUUGGUUAAAAA-A-------AAAGUGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUGCCCCUGAGCUAUACCCCCACUUAUGUU -......((((...(((......-.-------..(((...((((((((((((.......)))))((.(..(((((.....)))))..).)))))))))..)))..)))...))))..... ( -32.70) >DroSec_CAF1 7405 112 + 1 -AAUAACGAGUAUUGCUCAAAAAAA-------AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCACUUAAGUU -...((((((.....))).......-------........(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))......))) ( -32.30) >DroSim_CAF1 12671 110 + 1 -AAUAACGAGUAUUGCUCAAAAA---------AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCACUUAAGUU -...((((((.....))).....---------........(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))......))) ( -32.30) >DroEre_CAF1 13504 106 + 1 -AAUAACGAGUAUCCCCUAAAAA-------------AGAAAGGGGCACCCGGAUUCGAACCGGGGACCUCUCGAUCUGUGGUCGAAUGCUCUACUUCUGAGCUACACCCCCACUUGUGAG -....((((((............-------------.....((((..(((((.......)))))..))))(((((.....)))))..((((.......)))).........))))))... ( -32.10) >DroYak_CAF1 13405 106 + 1 -AAAAACGAGUAUCCCUUAAAAA-------------AGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCACUUGCCAC -.....(((((....(((....)-------------))..(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))))))).... ( -32.20) >DroAna_CAF1 17880 120 + 1 UUAUAAAGAAUACCAUUAAAGGAAAAAAAGCCAAAAAAAAGGGGGCACCCGGAUUUGAACCAGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUGAAAG ............((......))..................(((((..(((((.......)).))).....(((((.....)))))..((((.......))))....)))))......... ( -26.10) >consensus _AAUAACGAGUAUCGCUUAAAAA_________AAAAAGAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCACUUAAGAU ........................................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))......... (-27.12 = -27.45 + 0.33)



| Location | 21,787,904 – 21,788,015 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.33 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
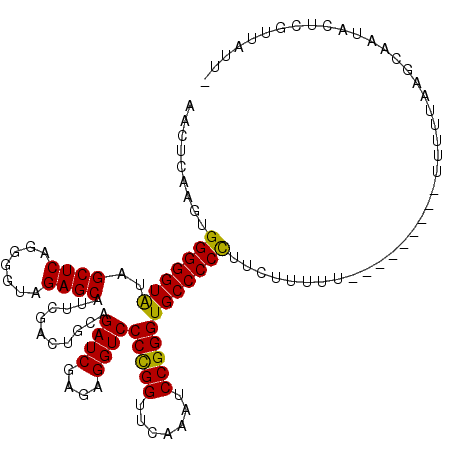
>3L_DroMel_CAF1 21787904 111 - 23771897 AACAUAAGUGGGGGUAUAGCUCAGGGGCAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCACUUU-------U-UUUUUAACCAAUACUCGUUAUU- .....(((((((((.....))).((((((..(((......))).((((....))))(((((.......)))))))))))..)))))).-------.-......................- ( -34.30) >DroSec_CAF1 7405 112 - 1 AACUUAAGUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU-------UUUUUUUGAGCAAUACUCGUUAUU- ...........((((((.(((((((((((..(((......))).((((....))))(((((.......))))))))))).........-------......))))).))))))......- ( -37.86) >DroSim_CAF1 12671 110 - 1 AACUUAAGUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU---------UUUUUGAGCAAUACUCGUUAUU- ...........((((((.(((((((((((..(((......))).((((....))))(((((.......))))))))))).........---------....))))).))))))......- ( -38.01) >DroEre_CAF1 13504 106 - 1 CUCACAAGUGGGGGUGUAGCUCAGAAGUAGAGCAUUCGACCACAGAUCGAGAGGUCCCCGGUUCGAAUCCGGGUGCCCCUUUCU-------------UUUUUAGGGGAUACUCGUUAUU- ((((....))))......((((.......))))((((((((...((((....))))...)).)))))).(((((((((((....-------------.....))))).)))))).....- ( -36.20) >DroYak_CAF1 13405 106 - 1 GUGGCAAGUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCU-------------UUUUUAAGGGAUACUCGUUUUU- ..(((.((((((((((..((((.......))))...........((((....))))(((((.......))))))))))))((((-------------(....)))))..))).)))...- ( -34.70) >DroAna_CAF1 17880 120 - 1 CUUUCAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCUGGUUCAAAUCCGGGUGCCCCCUUUUUUUUGGCUUUUUUUCCUUUAAUGGUAUUCUUUAUAA .........(((((((..((((.......))))...........((((....))))(((((.......))))))))))))..................((......))............ ( -31.00) >consensus AACUCAAGUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUCUUUUU_________UUUUUAAGCAAUACUCGUUAUU_ .........(((((((..((((.......))))...........((((....))))(((((.......))))))))))))........................................ (-31.75 = -31.33 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:30 2006