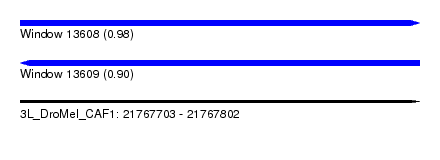
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,767,703 – 21,767,802 |
| Length | 99 |
| Max. P | 0.983588 |

| Location | 21,767,703 – 21,767,802 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
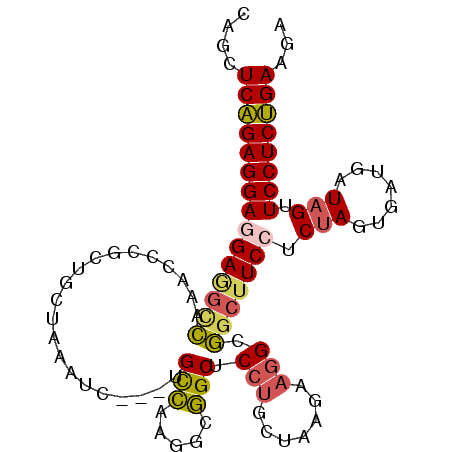
>3L_DroMel_CAF1 21767703 99 + 23771897 CAGCUCAGAGGAGGAGGUCAAACCCGCUGCUAAAUCUGCUGCCAAGUUGGCUCCUGCUAAGAAGGGGGCUUCCUCUAGUGAUGAUAGUUCCUCUGAAGA ....(((((((((((((((...(((((.((.......)).))....(((((....)))))...)))))))))).(((.......))).))))))))... ( -34.80) >DroSec_CAF1 2930 87 + 1 CACCUCAGAGGAGGAGGCCAAACCCGCU------------GCCAAGGCGGCUCCUGCUAAGAAGGCGGCUUCCCCUAGUGAUGAUAGUUCCUCUGAAGA ..(.(((((((((((((((......(((------------((....)))))....(((.....)))))))))).(((.......))).)))))))).). ( -37.40) >DroSim_CAF1 2446 87 + 1 CACCUCAGAGGAGGAGGCCAAACCCGCU------------GCCAAGGCGGCUCCUGCUAAGAAGGCGGCUUCCCCUAGUGAUGAUAGUUCCUCUGAAGA ..(.(((((((((((((((......(((------------((....)))))....(((.....)))))))))).(((.......))).)))))))).). ( -37.40) >DroEre_CAF1 2430 96 + 1 CAGCUCAGAGGAGGAGGCCAAGCCCGCAGCUAAAUC---UGCCAAGGCGGCUCCUGCUAAGAAGGCGGCUUCCUCUAGUGAUGAUAGUUCCUCUGAAGA ....(((((((((((((((..(((.((((......)---)))...)))(.((.((....)).)).)))))))).(((.......))).))))))))... ( -37.90) >DroYak_CAF1 2495 99 + 1 UAGCUCAGAGGAAGAGACCAAACCCUCUUCUAAAUCUUCUGUCAAGGCGGCUCCUGCUAAGAAGGCGGCGUCCUCUAGUGACGAUAGUUCCUCUGAAGA ....((((((((((((........)))........((..((((((((((.(.(((.......))).).)).)))....)))))..)))))))))))... ( -30.70) >DroPer_CAF1 3100 97 + 1 --ACUCCGAGGAAGAAGAAAAGCCGGCAGCAAAGGCUCCAGCUAAGGCAGCCCCCAAAAAGCUUGCUGCUUCCUCCAGCGAGGAUAGUUCCUCGGAGGA --.((((((((((...........((((((((.((((((......)).))))..........))))))))(((((....)))))...)))))))))).. ( -41.30) >consensus CAGCUCAGAGGAGGAGGCCAAACCCGCUGCUAAAUC___UGCCAAGGCGGCUCCUGCUAAGAAGGCGGCUUCCUCUAGUGAUGAUAGUUCCUCUGAAGA ....(((((((((((((((.....................(((.....))).(((.......))).))))))).(((.......))).))))))))... (-24.60 = -24.75 + 0.15)


| Location | 21,767,703 – 21,767,802 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
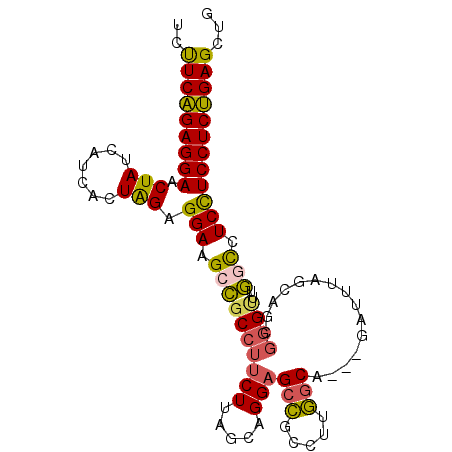
>3L_DroMel_CAF1 21767703 99 - 23771897 UCUUCAGAGGAACUAUCAUCACUAGAGGAAGCCCCCUUCUUAGCAGGAGCCAACUUGGCAGCAGAUUUAGCAGCGGGUUUGACCUCCUCCUCUGAGCUG ..(((((((((.............(((((((((((((.......))).(((.....))).((.......))...))))))..)))).)))))))))... ( -28.84) >DroSec_CAF1 2930 87 - 1 UCUUCAGAGGAACUAUCAUCACUAGGGGAAGCCGCCUUCUUAGCAGGAGCCGCCUUGGC------------AGCGGGUUUGGCCUCCUCCUCUGAGGUG .((((((((((.(((.......))).(((.((((((((((.....)))((.((....))------------.))))))..))).))))))))))))).. ( -33.60) >DroSim_CAF1 2446 87 - 1 UCUUCAGAGGAACUAUCAUCACUAGGGGAAGCCGCCUUCUUAGCAGGAGCCGCCUUGGC------------AGCGGGUUUGGCCUCCUCCUCUGAGGUG .((((((((((.(((.......))).(((.((((((((((.....)))((.((....))------------.))))))..))).))))))))))))).. ( -33.60) >DroEre_CAF1 2430 96 - 1 UCUUCAGAGGAACUAUCAUCACUAGAGGAAGCCGCCUUCUUAGCAGGAGCCGCCUUGGCA---GAUUUAGCUGCGGGCUUGGCCUCCUCCUCUGAGCUG ..(((((((((.(((.......))).(((.((((.(((((....))))).)((((..(((---(......))))))))..))).))))))))))))... ( -36.10) >DroYak_CAF1 2495 99 - 1 UCUUCAGAGGAACUAUCGUCACUAGAGGACGCCGCCUUCUUAGCAGGAGCCGCCUUGACAGAAGAUUUAGAAGAGGGUUUGGUCUCUUCCUCUGAGCUA ..((((((((((.....(((.(....))))(((((((((((..((((......)))).............))))))))..)))...))))))))))... ( -28.86) >DroPer_CAF1 3100 97 - 1 UCCUCCGAGGAACUAUCCUCGCUGGAGGAAGCAGCAAGCUUUUUGGGGGCUGCCUUAGCUGGAGCCUUUGCUGCCGGCUUUUCUUCUUCCUCGGAGU-- ..((((((((((...(((((....))))).(((((((.((....))(((((.((......))))))))))))))............)))))))))).-- ( -43.60) >consensus UCUUCAGAGGAACUAUCAUCACUAGAGGAAGCCGCCUUCUUAGCAGGAGCCGCCUUGGCA___GAUUUAGCAGCGGGUUUGGCCUCCUCCUCUGAGCUG ..(((((((((.(((.......))).(((.((((((((((.....)))(((.....)))...............))))..))).))))))))))))... (-23.60 = -23.13 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:23 2006