| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,530,754 – 2,530,856 |
| Length | 102 |
| Max. P | 0.661772 |

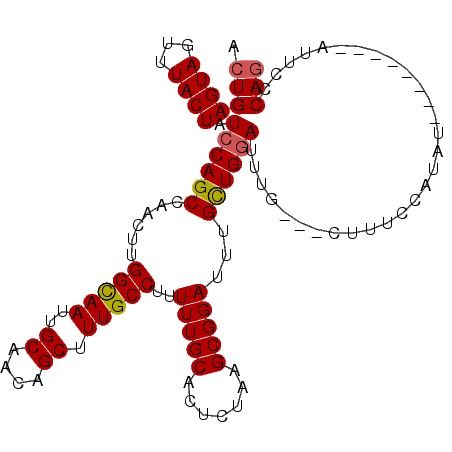
| Location | 2,530,754 – 2,530,856 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2530754 102 + 23771897 CUGGGAAU--------AUAUGGAAAGCAGCAAAUCCAGCAAAUCCGCUUAGAGUGCAAAAAGGCAAAGCUGUUGCAAUUGCCAAGUUGGCUGGAUAGUAAACUACUCAUU .((((...--------.................((((((.....(((.....)))(((...(((((.((....))..)))))...)))))))))(((....))))))).. ( -23.60) >DroSec_CAF1 153602 99 + 1 CUGGGAAU--------AUAUGGAAAC---CAAAUCCAGCAAAUCCGCUUAGAGUGCAAAAAGGCAAAGCUGUUGCAAUUACCAAGUUGGCUGGAUAGUAAACUACUCAGC (((((...--------....(....)---...(((((((......((((.(..(((((...(((...))).)))))....).))))..))))))).........))))). ( -25.00) >DroSim_CAF1 151160 99 + 1 CUGGGAAU--------AUAUGGAAAG---CAAAUCCAGCAAAUCCGCUUAGAGUGCAAAAAGGCAAAGCUGUUGCAAUUGCCAAGUUGGCUGGAUAGUAAACUACUCAGU (((((...--------..........---....((((((.....(((.....)))(((...(((((.((....))..)))))...)))))))))(((....)))))))). ( -26.10) >DroYak_CAF1 153691 107 + 1 CUGGGAAUAUGUAUAUAUAUGCAAAG---CAAAUGCAACAAAUCCGCUUAGAGUGCAAAAAGGCAAAGCUGUGGCAAUUGCCAAGCUGGCUGGAGAGUAAACUACUCAGU (((((....(((((....)))))..(---(....)).((...(((((.......)).....(((..((((.((((....)))))))).))))))..))......))))). ( -27.00) >consensus CUGGGAAU________AUAUGGAAAG___CAAAUCCAGCAAAUCCGCUUAGAGUGCAAAAAGGCAAAGCUGUUGCAAUUGCCAAGUUGGCUGGAUAGUAAACUACUCAGU (((((..............((((..........))))....((((((.(((..(((......)))...)))..))....(((.....))).)))).........))))). (-19.52 = -19.90 + 0.38)

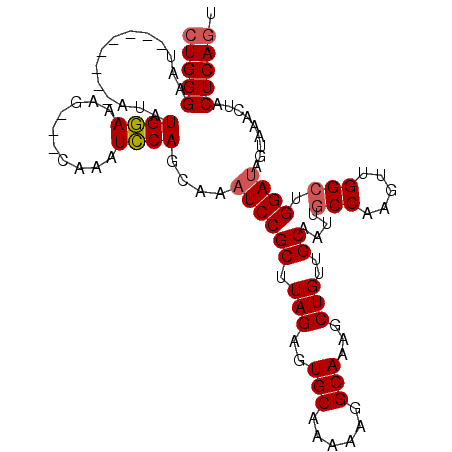
| Location | 2,530,754 – 2,530,856 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2530754 102 - 23771897 AAUGAGUAGUUUACUAUCCAGCCAACUUGGCAAUUGCAACAGCUUUGCCUUUUUGCACUCUAAGCGGAUUUGCUGGAUUUGCUGCUUUCCAUAU--------AUUCCCAG ...(((((((.....(((((((......(((((..((....)).)))))..(((((.......)))))...)))))))..))))))).......--------........ ( -28.90) >DroSec_CAF1 153602 99 - 1 GCUGAGUAGUUUACUAUCCAGCCAACUUGGUAAUUGCAACAGCUUUGCCUUUUUGCACUCUAAGCGGAUUUGCUGGAUUUG---GUUUCCAUAU--------AUUCCCAG ...(((((....((((((((((......(((((..((....)).)))))..(((((.......)))))...)))))))..)---)).......)--------)))).... ( -22.60) >DroSim_CAF1 151160 99 - 1 ACUGAGUAGUUUACUAUCCAGCCAACUUGGCAAUUGCAACAGCUUUGCCUUUUUGCACUCUAAGCGGAUUUGCUGGAUUUG---CUUUCCAUAU--------AUUCCCAG ...(((.(((.....(((((((......(((((..((....)).)))))..(((((.......)))))...)))))))..)---))))).....--------........ ( -23.50) >DroYak_CAF1 153691 107 - 1 ACUGAGUAGUUUACUCUCCAGCCAGCUUGGCAAUUGCCACAGCUUUGCCUUUUUGCACUCUAAGCGGAUUUGUUGCAUUUG---CUUUGCAUAUAUAUACAUAUUCCCAG ...(((((...)))))........((..(((((.(((.((((.(((((...............))))).)))).))).)))---))..)).................... ( -23.56) >consensus ACUGAGUAGUUUACUAUCCAGCCAACUUGGCAAUUGCAACAGCUUUGCCUUUUUGCACUCUAAGCGGAUUUGCUGGAUUUG___CUUUCCAUAU________AUUCCCAG .(((((((...)))).((((((......(((((..((....)).)))))..(((((.......)))))...))))))..............................))) (-18.27 = -18.40 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:14 2006