
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,748,744 – 21,748,904 |
| Length | 160 |
| Max. P | 0.984450 |

| Location | 21,748,744 – 21,748,864 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748744 120 + 23771897 AUAAUUCAUUUGUGACAAUUGUGAGAAACUGUAGAGAUGCGAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCGAAAUCGAA .....((((((.............((((.((((....)))).))))((((((((.(....).))))))))...))))))..((((((((..(((((..........))))))))))))). ( -28.00) >DroSec_CAF1 34007 120 + 1 AGAAUUCAUUUGUGACAAUUGUGAGAAACUGUAGAGAUGCGAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAA .....((((((.............((((.((((....)))).))))((((((((.(....).))))))))...))))))..(((((((...(((((..........))))).))))))). ( -27.70) >DroSim_CAF1 35891 120 + 1 AGAAUUCAUUUGUGACAAUUGUGAGAAACUGUAGAGAUGCGAUUUCGCACAUUUAGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAA .....((((((.............((((.((((....)))).))))((((((((.(....).))))))))...))))))..(((((((...(((((..........))))).))))))). ( -27.70) >DroEre_CAF1 16526 119 + 1 AUAACUCAUUUGUGACA-UUGUGAGAAACUGUUGGGAUGCGAUUUCGCACAUUUCGUAGACAAAAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAA .....((((((......-......((((.(((......))).))))((((((((.(....).))))))))...))))))..(((((((...(((((..........))))).))))))). ( -26.40) >consensus AGAAUUCAUUUGUGACAAUUGUGAGAAACUGUAGAGAUGCGAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAA .....((((((.............((((.((((....)))).))))((((((((.(....).))))))))...))))))..((((((((..(((((..........))))))))))))). (-27.25 = -27.12 + -0.12)

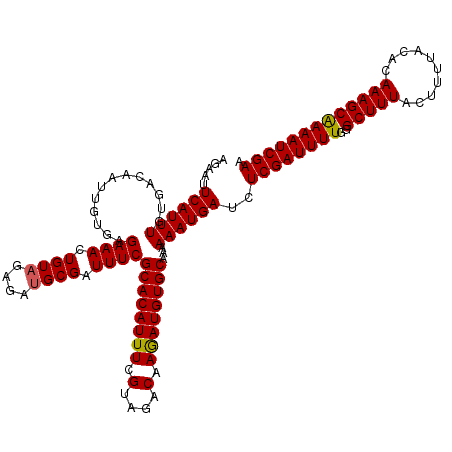
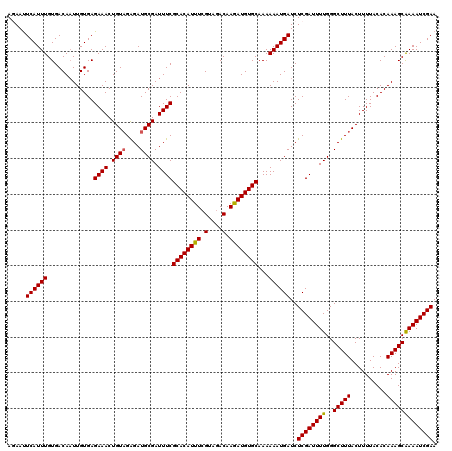
| Location | 21,748,744 – 21,748,864 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -26.88 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748744 120 - 23771897 UUCGAUUUCGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUCGCAUCUCUACAGUUUCUCACAAUUGUCACAAAUGAAUUAU ((((((((.((((((........))))))...)))))))).(((((((((((((((.(((....))).)))))))))...........((((((......))))))...))))))..... ( -28.00) >DroSec_CAF1 34007 120 - 1 UUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUCGCAUCUCUACAGUUUCUCACAAUUGUCACAAAUGAAUUCU (((((((((((((((........))))))..))))))))).(((((((((((((((.(((....))).)))))))))...........((((((......))))))...))))))..... ( -29.00) >DroSim_CAF1 35891 120 - 1 UUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACUAAAUGUGCGAAAUCGCAUCUCUACAGUUUCUCACAAUUGUCACAAAUGAAUUCU (((((((((((((((........))))))..))))))))).(((((((((((((((...(....)...)))))))))...........((((((......))))))...))))))..... ( -26.00) >DroEre_CAF1 16526 119 - 1 UUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUUUUGUCUACGAAAUGUGCGAAAUCGCAUCCCAACAGUUUCUCACAA-UGUCACAAAUGAGUUAU (((((((((((((((........))))))..))))))))).((((((...((((((((((....))))))))))(((((.(........).)))))......-......))))))..... ( -29.90) >consensus UUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUCGCAUCUCUACAGUUUCUCACAAUUGUCACAAAUGAAUUAU (((((((((((((((........))))))..))))))))).(((((((((((((((.(((....))).)))))))))...........((((((......))))))...))))))..... (-26.88 = -27.62 + 0.75)

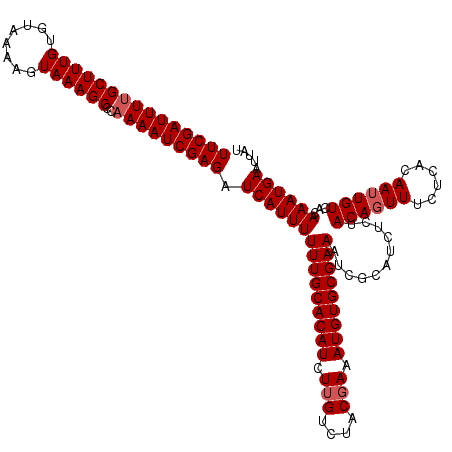
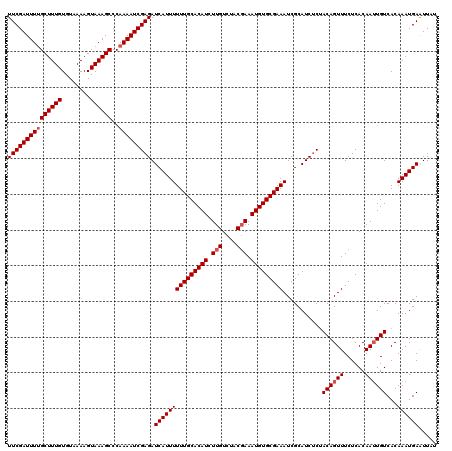
| Location | 21,748,784 – 21,748,904 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748784 120 + 23771897 GAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCGAAAUCGAAAACAUUAUUACUGUAAUUAAAAUUCAAUUCGAAAAAAUAA ..((((((((((((.(....).))))))))....((((...((((((((..(((((..........)))))))))))))...))))........................))))...... ( -23.30) >DroSec_CAF1 34047 120 + 1 GAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAAAACAUUAUUACUGUAAUUAAAAUUCAAUUCGGUAAAUUAA ......((((((((.(....).))))))))....((((...(((((((...(((((..........))))).)))))))...)))).((((((.((((.......))))))))))..... ( -25.20) >DroSim_CAF1 35931 120 + 1 GAUUUCGCACAUUUAGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAAAACAUUAUUACUGUAAUUAAAAUUCAAUUCGGUAAAUUAA ......((((((((.(....).))))))))....((((...(((((((...(((((..........))))).)))))))...)))).((((((.((((.......))))))))))..... ( -25.20) >DroEre_CAF1 16565 120 + 1 GAUUUCGCACAUUUCGUAGACAAAAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAAAACAUUAUUACUGUAAUUAAAAUUCAAUUCGGUAAAUUAA ......((((((((.(....).))))))))....((((...(((((((...(((((..........))))).)))))))...)))).((((((.((((.......))))))))))..... ( -25.10) >consensus GAUUUCGCACAUUUCGUAGACAAGAUGUGCAAAAAAUGAUCUCGAUUUUGGGCUUUACUUUUACACAAAGCAAAAUCGAAAACAUUAUUACUGUAAUUAAAAUUCAAUUCGGUAAAUUAA ......((((((((.(....).))))))))....((((...((((((((..(((((..........)))))))))))))...)))).((((((.((((.......))))))))))..... (-23.65 = -23.77 + 0.13)


| Location | 21,748,784 – 21,748,904 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -27.99 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748784 120 - 23771897 UUAUUUUUUCGAAUUGAAUUUUAAUUACAGUAAUAAUGUUUUCGAUUUCGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUC ..................................((((.(((((((((.((((((........))))))...))))))))).))))((((((((((.(((....))).)))))))))).. ( -27.60) >DroSec_CAF1 34047 120 - 1 UUAAUUUACCGAAUUGAAUUUUAAUUACAGUAAUAAUGUUUUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUC .....((((.(((((((...)))))).).)))).((((.((((((((((((((((........))))))..)))))))))).))))((((((((((.(((....))).)))))))))).. ( -30.50) >DroSim_CAF1 35931 120 - 1 UUAAUUUACCGAAUUGAAUUUUAAUUACAGUAAUAAUGUUUUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACUAAAUGUGCGAAAUC .....((((.(((((((...)))))).).)))).((((.((((((((((((((((........))))))..)))))))))).))))((((((((((...(....)...)))))))))).. ( -27.50) >DroEre_CAF1 16565 120 - 1 UUAAUUUACCGAAUUGAAUUUUAAUUACAGUAAUAAUGUUUUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUUUUGUCUACGAAAUGUGCGAAAUC .....((((.(((((((...)))))).).)))).((((.((((((((((((((((........))))))..)))))))))).))))((((((((((((((....)))))))))))))).. ( -34.00) >consensus UUAAUUUACCGAAUUGAAUUUUAAUUACAGUAAUAAUGUUUUCGAUUUUGCUUUGUGUAAAAGUAAAGCCCAAAAUCGAGAUCAUUUUUUGCACAUCUUGUCUACGAAAUGUGCGAAAUC .....((((.(((((((...)))))).).)))).((((.((((((((((((((((........))))))..)))))))))).))))((((((((((.(((....))).)))))))))).. (-27.99 = -28.55 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:10 2006