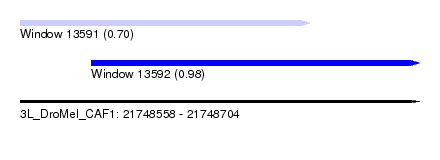
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,748,558 – 21,748,704 |
| Length | 146 |
| Max. P | 0.979699 |

| Location | 21,748,558 – 21,748,664 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748558 106 + 23771897 AAGCACAUUUU-----GGUUCAUUUAAUA------UCAAAUGGAUUAUUAACUUAUGAUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUU ..(((((((((-----(((((((((....------.((((.(((((((......))))))).))))....))))))))).((...)))))))))))(((.((((...)))).))).. ( -37.30) >DroSec_CAF1 33810 117 + 1 ACGCACUUUUUAUUUGAAUUUGUUCAAUAUUUCAAUCAAAUGGAUUAUUAAUUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUU ..(((........(((((....))))).........((((.(((..(.....)..)))....)))).)))....((((((((((((((.....)))((....)).))))))))))). ( -27.40) >DroSim_CAF1 35698 113 + 1 ----ACUUUUUAUUUGAAUUCGUUCAAUAUUUGAAUCAAAUGGAUUAUUAACUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUU ----.........(((((....)))))((((((...((((.(((...........)))....))))...))))))(((((((((((((.....)))((....)).)))))))))).. ( -28.80) >consensus A_GCACUUUUUAUUUGAAUUCGUUCAAUAUUU_AAUCAAAUGGAUUAUUAACUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUU ................((((((((..((......))..))))))))............................((((((((((((((.....)))((....)).))))))))))). (-23.55 = -23.00 + -0.55)



| Location | 21,748,584 – 21,748,704 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21748584 120 + 23771897 AAAUGGAUUAUUAACUUAUGAUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGUGAGUU (((.(((((((......))))))).))).........((((((((((((((.....)))((....)).)))))))))))((((((((.(((.((((....)))).))).))))))))... ( -40.80) >DroSec_CAF1 33847 120 + 1 AAAUGGAUUAUUAAUUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGUGAGUU ....(((..(.....)..)))................((((((((((((((.....)))((....)).)))))))))))((((((((.(((.((((....)))).))).))))))))... ( -36.90) >DroSim_CAF1 35731 120 + 1 AAAUGGAUUAUUAACUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGUGAGUU ....(((...........)))................((((((((((((((.....)))((....)).)))))))))))((((((((.(((.((((....)))).))).))))))))... ( -36.70) >DroEre_CAF1 16370 116 + 1 ----UUACUAUUAAUUUCUCCUCAAUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGUGAGUU ----................(((((....))).))..((((((((((((((.....)))((....)).)))))))))))((((((((.(((.((((....)))).))).))))))))... ( -36.20) >DroYak_CAF1 93385 115 + 1 AAGUGCACUCUUUAUUUCUCCUUUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGU----- ...((((..((((((............((((.((((.((((((((((((((.....)))((....)).)))))))))))....)))).)))).......))))))..))))....----- ( -37.11) >consensus AAAUGGAUUAUUAAUUUCUCCUCUUUUUGUUGCAGAUGAACCAGCUGUGCAAAGUGUGCGGCGAGCCGGCGGCUGGUUUUCAUUUUGGGGCAUUUACAUGUGAAGGCUGCAAGGUGAGUU .....................................((((((((((((((.....)))((....)).)))))))))))((((((((.(((.((((....)))).))).))))))))... (-34.60 = -35.00 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:12:06 2006