
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
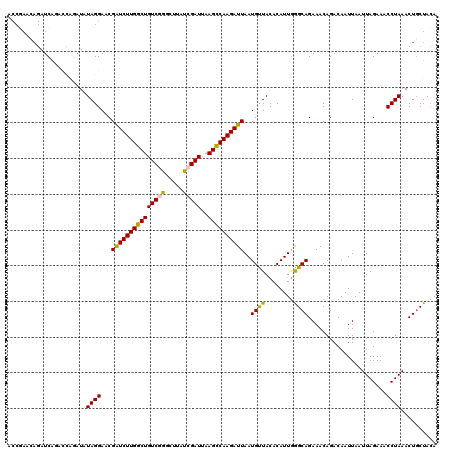
| Location | 21,744,545 – 21,744,676 |
| Length | 131 |
| Max. P | 0.979019 |

| Location | 21,744,545 – 21,744,665 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -22.44 |
| Energy contribution | -21.88 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744545 120 - 23771897 ACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCAGACUUAUCGAUUAAGCCAAGAUUAAUGUUACACAUUGGGCAGAAACAGACAAUUAAUUAGAAACCUAAACUGCUACA ......((((((......))).((((...(((((((((((((.((....)))))..))))))))))((((.....))))...........................))))..)))..... ( -22.10) >DroSec_CAF1 26308 120 - 1 ACCGAACAGAUCAGACCAGAUAUAGGAACGGUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGCUACACAUUGGGCAGAAACAGACAAUUAAUUAGAAACCUAAACUUCUACA .................(((..((((...(((((((((((((((.....)))))..))))))))))..((((........))))......................))))....)))... ( -26.10) >DroSim_CAF1 31189 120 - 1 ACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGCUACACAUUGGGCAGAAACAGACAAUUAAUUAGAAACCUAAACUGCUACA ......((((((......))).((((...(((((((((((((((.....)))))..))))))))))..((((........))))......................))))..)))..... ( -28.20) >DroEre_CAF1 12640 116 - 1 ACCAAACAGAUCACA----AUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGUUACACAUUGAGCAGAAACAGACAAUUAAUUAGAAACCUAAACUGCUACA .............((----((.....((((((((((((((((((.....)))))..))))))))))...)))....))))(((((((........))..((((....)))).)))))... ( -24.10) >DroYak_CAF1 84460 106 - 1 ----------UCGCA----AUAUAGGAACGAUCUUGGCUGUCAAUUUUAUCGAUUAAGUCAAGAUUAAUGUUACACAUUAAGCAGAAACAGACAAUUAAUUAGAAACCUAAACUGCUAAA ----------..(((----.(.((((...(((((((((((((.........)))..))))))))))((((.....))))...........................)))).).))).... ( -19.70) >consensus ACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGUUACACAUUGGGCAGAAACAGACAAUUAAUUAGAAACCUAAACUGCUACA ......................((((...(((((((((((((((.....)))))..))))))))))..((((........))))......................)))).......... (-22.44 = -21.88 + -0.56)


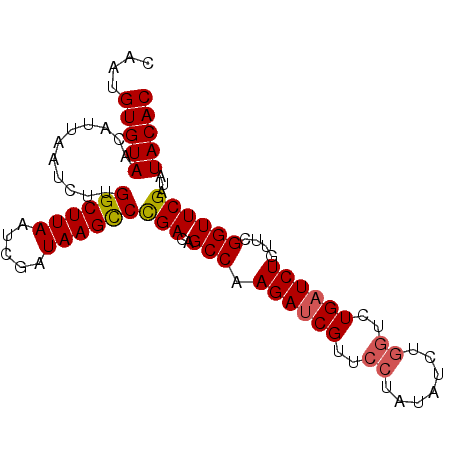
| Location | 21,744,585 – 21,744,676 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744585 91 + 23771897 CAAUGUGUAACAUUAAUCUUGGCUUAAUCGAUAAGUCUGACAGCCAAGAUCGUUCCUAUAUCUGGUCUGAUCUGUUCGGUUCGAUAUACAC ....(((((......(((((((((...(((.......))).)))))))))........((((.(..((((.....))))..)))))))))) ( -21.10) >DroSec_CAF1 26348 91 + 1 CAAUGUGUAGCAUUAAUCUUGGCUUAAUCGAUAAGCCCGACAGCCAAGACCGUUCCUAUAUCUGGUCUGAUCUGUUCGGUUCGAUGUACAC ....(((((.((((((((..((((((.....)))))).(((((...((((((..........))))))...))))).)))).))))))))) ( -26.30) >DroSim_CAF1 31229 91 + 1 CAAUGUGUAGCAUUAAUCUUGGCUUAAUCGAUAAGCCCGACAGCCAAGAUCGUUCCUAUAUCUGGUCUGAUCUGUUCGGUUCGAUGUACAC ....(((((.((((((((.(((((...(((.......))).)))))((((((..((.......))..))))))....)))).))))))))) ( -24.80) >DroEre_CAF1 12680 87 + 1 CAAUGUGUAACAUUAAUCUUGGCUUAAUCGAUAAGCCCGACAGCCAAGAUCGUUCCUAUAU----UGUGAUCUGUUUGGUUCGAUAUACAC ...((((((.......((((((((...(((.......))).))))))))(((..((.(((.----.......)))..))..))))))))). ( -21.40) >consensus CAAUGUGUAACAUUAAUCUUGGCUUAAUCGAUAAGCCCGACAGCCAAGAUCGUUCCUAUAUCUGGUCUGAUCUGUUCGGUUCGAUAUACAC ....(((((...........((((((.....))))))(((..(((.((((((..((.......))..))))))....))))))...))))) (-19.25 = -19.62 + 0.38)


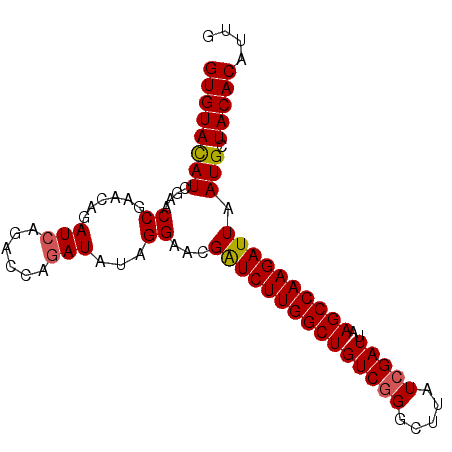
| Location | 21,744,585 – 21,744,676 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744585 91 - 23771897 GUGUAUAUCGAACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCAGACUUAUCGAUUAAGCCAAGAUUAAUGUUACACAUUG (((((.......((......(((......)))...))...(((((((((((((.((....)))))..)))))))))).....))))).... ( -22.00) >DroSec_CAF1 26348 91 - 1 GUGUACAUCGAACCGAACAGAUCAGACCAGAUAUAGGAACGGUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGCUACACAUUG ((((((((..(..(((.(((.(((((((............)))).)))))))))((((((.....))))))....)..))).))))).... ( -28.10) >DroSim_CAF1 31229 91 - 1 GUGUACAUCGAACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGCUACACAUUG ((((((((....((......(((......)))...))...(((((((((((((((.....)))))..)))))))))).))).))))).... ( -26.90) >DroEre_CAF1 12680 87 - 1 GUGUAUAUCGAACCAAACAGAUCACA----AUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGUUACACAUUG (((((.......((............----.....))...(((((((((((((((.....)))))..)))))))))).....))))).... ( -22.93) >consensus GUGUACAUCGAACCGAACAGAUCAGACCAGAUAUAGGAACGAUCUUGGCUGUCGGGCUUAUCGAUUAAGCCAAGAUUAAUGCUACACAUUG ((((((((....((......(((......)))...))...(((((((((((((((.....)))))..)))))))))).))).))))).... (-23.26 = -23.32 + 0.06)

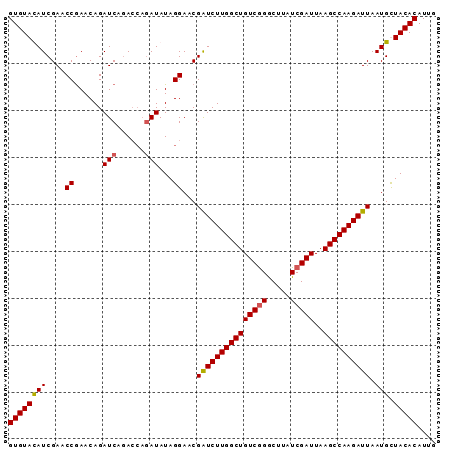
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:57 2006