
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,744,109 – 21,744,465 |
| Length | 356 |
| Max. P | 0.938483 |

| Location | 21,744,109 – 21,744,227 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744109 118 - 23771897 AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUCAAGAAC--ACAGCCCACUGAACCGAACCCCGGAGCAUUGACACAAUCUGCAAUCCAAAUC .....(((......((((((.((((.(((((((.((...)).))))))).................--.(((....)))..(((.....)))....)))).))))))))).......... ( -18.10) >DroSec_CAF1 25875 118 - 1 AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUCAAGAAC--ACAGCCCACUGAACCGAACCCCGGAGCAUUGACACAAUCUGCAAUCCAAAUC .....(((......((((((.((((.(((((((.((...)).))))))).................--.(((....)))..(((.....)))....)))).))))))))).......... ( -18.10) >DroSim_CAF1 30756 118 - 1 AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUCAAGAAC--ACAGCCCACUGAACCGAACCCCGGAGCAUUGACACAAUCUGCAAUCCAAAUC .....(((......((((((.((((.(((((((.((...)).))))))).................--.(((....)))..(((.....)))....)))).))))))))).......... ( -18.10) >DroEre_CAF1 12217 114 - 1 AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUGCAGAACACACAGCCCACUGA-----ACCCCGGAGCAUUGACACAAUCUGCA-UCCAAAUC .....(((......((((((.((...(((((((.((...)).))))))).......(((((........(((....))).-----.(....).))))))).))))))))).-........ ( -17.70) >DroYak_CAF1 84041 111 - 1 AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUCAAGAAC--ACAGCCCACUGA-----ACCCCGGAGCAUUGACACAAUCUGCAAUCCAAA-- .....(((......((((((.((((.(((((((.((...)).))))))).................--...((((.....-----.....)).)).)))).)))))))))........-- ( -17.10) >consensus AACAAGCAAACCACGAUUGUCUCAAAUAUUUUGAAGCCGCUACAAAAUAGAACAAGAAUCAAGAAC__ACAGCCCACUGAACCGAACCCCGGAGCAUUGACACAAUCUGCAAUCCAAAUC .....(((......((((((.((((.(((((((.((...)).)))))))......................((((...............)).)).)))).))))))))).......... (-16.66 = -16.66 + 0.00)

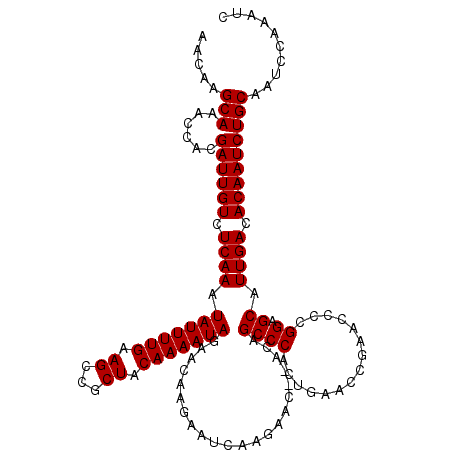
| Location | 21,744,227 – 21,744,345 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -19.01 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744227 118 - 23771897 UGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--AAAAAAUGUGUCCUUUAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCUGAUAAAGGCAACAGAGGGAGAAAAGGUGUUCA .(((((((((((....(((.............)))..--......(((((((.(((............))).))))..((((............)))))))..))))))).....)))). ( -20.52) >DroSec_CAF1 25993 117 - 1 UGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--AA-AAAUGUUUCCUUUAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCUGAUAAAGGCAACAGAGGGAGAAAAGGUGUUCA .(((((((((((...................((((..--..-...(((((.((....)).))))).......))))..((((............)))).....))))))).....)))). ( -22.94) >DroSim_CAF1 30874 117 - 1 UGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--AA-AAAUGUGUCCUUUAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCUGAUAAAGGCAACAGAGGGAGAAAAGGUGUUCA .(((((((((((...................((((..--..-...(((...((....))...))).......))))..((((............)))).....))))))).....)))). ( -20.64) >DroEre_CAF1 12331 119 - 1 UGGAUUUCUCUCAAUAAGAAUAAUAAACAAGGUUCCAAAAA-AAAUGUGUCCUUAAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCAGAUAAAGGCAACAGAGGGAGAAAAGGUGUUCA ....((((((((.............................-.....(((((....................))))).((((............))))...))))))))........... ( -19.75) >DroYak_CAF1 84152 117 - 1 UGGAUUUCUCUCAAUAAGAAUAAUAAACAAGGUU--AAAAA-AAAUGUGUCCUUAAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCAGAUAAAGGCAACACAGGGAGAAAAGGUGUUCA ....((((((((......................--.....-.....(((((....................))))).((((............)))).....))))))))......... ( -19.65) >consensus UGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA__AA_AAAUGUGUCCUUUAAAGCAAACAAAAUGAUGGACAAUGCCAAAUCUGAUAAAGGCAACAGAGGGAGAAAAGGUGUUCA .(((((((((((...................................(((((....................))))).((((............)))).....))))))).....)))). (-19.01 = -18.97 + -0.04)

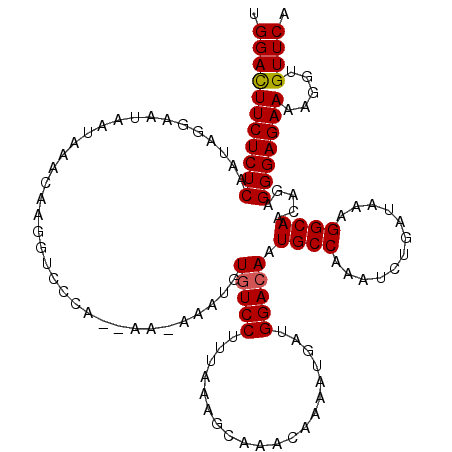

| Location | 21,744,307 – 21,744,425 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.29 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744307 118 + 23771897 U--UGGGACCUUGUUUAUUAUUCCUAUUGAGAGAAGUCCAACGUUCUGCCCACUUUUUUUCAGCUCUGUCAACAGCGAGUUGAUGAGCUUGUUGAGAAUGUGGCAGCGACCAACUGAUCU .--..((((.((.((((.((....)).)))).)).))))...((((((((((.(((((..((((((.((((((.....)))))))))).))..))))))).))))).))).......... ( -32.50) >DroSec_CAF1 26072 118 + 1 U--UGGGACCUUGUUUAUUAUUCCUAUUGAGAGAAGUCCAACGUUCUGCCCAUUUUUUUCCAGCUCUGUCAACAGCGACUUGAUGAGCUUGUUGAGAAUGUGGCAGCGACCAACUGAUCU .--..((((.((.((((.((....)).)))).)).))))...(((((((((((((((..(.(((((.(((((.......)))))))))).)..))))))).))))).))).......... ( -33.10) >DroSim_CAF1 30953 118 + 1 U--UGGGACCUUGUUUAUUAUUCCUAUUGAGAGAAGUCCAACGUUCUGCCCAUUUUUUUCCAGCUCUGUCAACAGCGAGUUGAUGAGCUUGUUGAGAAUGUGGCAGCGACCAACUGAUCU .--..((((.((.((((.((....)).)))).)).))))...(((((((((((((((..(.(((((.((((((.....))))))))))).)..))))))).))))).))).......... ( -36.90) >DroEre_CAF1 12410 119 + 1 UUUUGGAACCUUGUUUAUUAUUCUUAUUGAGAGAAAUCCAACGUUCUUCACAUUUUUU-UCAGCUCUGUCAACAGCGAGUUGAUGAGCUUGUUGAGAAUGUGGCAGCGACCAACUGAUCU ..((((....................(((.((....)))))((((..((((((((((.-.((((((.((((((.....)))))))))).))..)))))))))).)))).))))....... ( -33.20) >DroYak_CAF1 84231 117 + 1 UUUU--AACCUUGUUUAUUAUUCUUAUUGAGAGAAAUCCAACGUUCUUCCCAUUUAUU-UCAGCUCUGUCAACAGAGAGUUGAUGAGCUUGUUGAGAAUGUGGCAGCUACCAACUGAUCU ....--..............(((((.....)))))..(((.(((((((..((......-..(((((.((((((.....)))))))))))))..))))))))))(((.......))).... ( -25.00) >consensus U__UGGGACCUUGUUUAUUAUUCCUAUUGAGAGAAGUCCAACGUUCUGCCCAUUUUUUUUCAGCUCUGUCAACAGCGAGUUGAUGAGCUUGUUGAGAAUGUGGCAGCGACCAACUGAUCU ...((((((.(((..(.((.(((.....))).)).)..))).)))((((((((((((..(.(((((.((((((.....))))))))))).)..))))))).)))))...)))........ (-25.02 = -25.90 + 0.88)

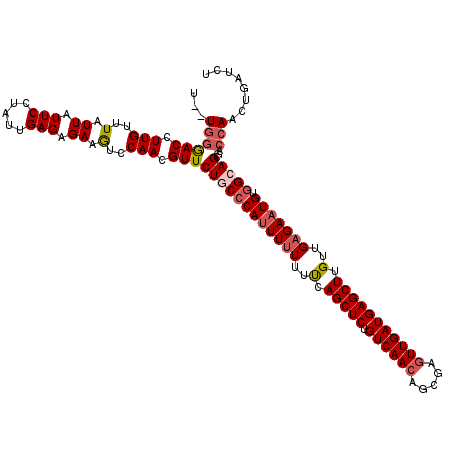
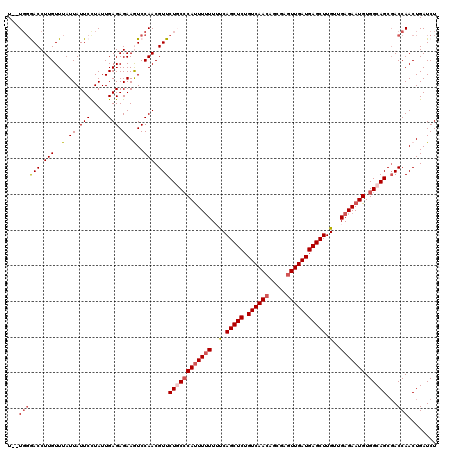
| Location | 21,744,307 – 21,744,425 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.29 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744307 118 - 23771897 AGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGAAAAAAAGUGGGCAGAACGUUGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--A .((((....)))).(((((.((((.......(((((.(((((.....)))))..))))).......)))))))))......((((((......................))))))..--. ( -31.39) >DroSec_CAF1 26072 118 - 1 AGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAAGUCGCUGUUGACAGAGCUGGAAAAAAAUGGGCAGAACGUUGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--A .((((....)))).(((((.((((.......(((((.((((.......))))..))))).......)))))))))......((((((......................))))))..--. ( -27.09) >DroSim_CAF1 30953 118 - 1 AGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGGAAAAAAAUGGGCAGAACGUUGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA--A .((((....)))).(((((.((((.......(((((.(((((.....)))))..))))).......)))))))))......((((((......................))))))..--. ( -31.39) >DroEre_CAF1 12410 119 - 1 AGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGA-AAAAAAUGUGAAGAACGUUGGAUUUCUCUCAAUAAGAAUAAUAAACAAGGUUCCAAAA .((((....)))).....((((((.......(((((.(((((.....)))))..)))))..-....))))))..((((.(((.(((..(((.....)))..)))...))).))))..... ( -29.32) >DroYak_CAF1 84231 117 - 1 AGAUCAGUUGGUAGCUGCCACAUUCUCAACAAGCUCAUCAACUCUCUGUUGACAGAGCUGA-AAUAAAUGGGAAGAACGUUGGAUUUCUCUCAAUAAGAAUAAUAAACAAGGUU--AAAA ......((.(((....))))).((((((...(((((.(((((.....)))))..)))))..-......)))))).(((.(((.(((..(((.....)))..)))...))).)))--.... ( -24.20) >consensus AGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGAAAAAAAAUGGGCAGAACGUUGGACUUCUCUCAAUAGGAAUAAUAAACAAGGUCCCA__A .((((.....((..(((((.((((.......(((((.(((((.....)))))..))))).......))))))))).))(((....((((.......)))).....)))..))))...... (-24.00 = -24.16 + 0.16)

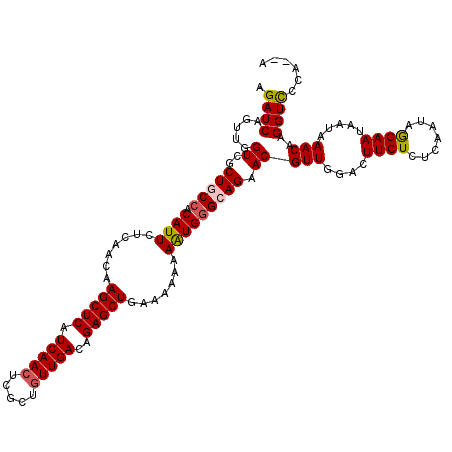
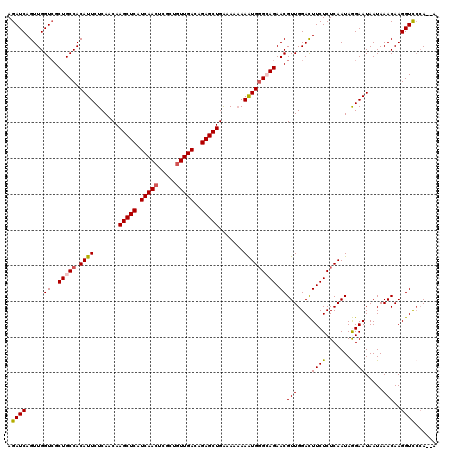
| Location | 21,744,345 – 21,744,465 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -28.21 |
| Energy contribution | -29.45 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21744345 120 - 23771897 CGAUGGGGACAAGUUCAGAGCGGCUUAGACAAUAAGAACCAGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGAAAAAAAGUGGGCAGAACGU ...((....)).((((..((((.((((.....)))).(((((.....)))))))))(((.((((.......(((((.(((((.....)))))..))))).......))))))).)))).. ( -36.34) >DroSec_CAF1 26110 120 - 1 CGAUGGGGACAAGUUCAGAGCGGCUUAGACAAUAAGAACCAGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAAGUCGCUGUUGACAGAGCUGGAAAAAAAUGGGCAGAACGU ...((....)).((((..((((.((((.....)))).(((((.....)))))))))(((.((((.......(((((.((((.......))))..))))).......))))))).)))).. ( -32.04) >DroSim_CAF1 30991 120 - 1 CGAUGGGGACAAGUUCAGAGCGGCUUAGACAAUAAGAACCAGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGGAAAAAAAUGGGCAGAACGU ...((....)).((((..((((.((((.....)))).(((((.....)))))))))(((.((((.......(((((.(((((.....)))))..))))).......))))))).)))).. ( -36.34) >DroEre_CAF1 12450 118 - 1 CGG-GGGGACAAGUUCAGAGCGGCUUAGACAAUAAAAACCAGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGA-AAAAAAUGUGAAGAACGU ...-(....)..((((...((((((((.....)))......((((....)))))))))((((((.......(((((.(((((.....)))))..)))))..-....))))))..)))).. ( -33.22) >DroYak_CAF1 84269 119 - 1 UGAUGGGGACAAGUACAGAGCGGCUUAGACAAUAAGAACCAGAUCAGUUGGUAGCUGCCACAUUCUCAACAAGCUCAUCAACUCUCUGUUGACAGAGCUGA-AAUAAAUGGGAAGAACGU ...((....)).((.(.(.((((((............(((((.....))))))))))))...((((((...(((((.(((((.....)))))..)))))..-......))))))).)).. ( -28.70) >consensus CGAUGGGGACAAGUUCAGAGCGGCUUAGACAAUAAGAACCAGAUCAGUUGGUCGCUGCCACAUUCUCAACAAGCUCAUCAACUCGCUGUUGACAGAGCUGAAAAAAAAUGGGCAGAACGU ...((....)).((((..(((((((..(((................))))))))))(((.((((.......(((((.(((((.....)))))..))))).......))))))).)))).. (-28.21 = -29.45 + 1.24)

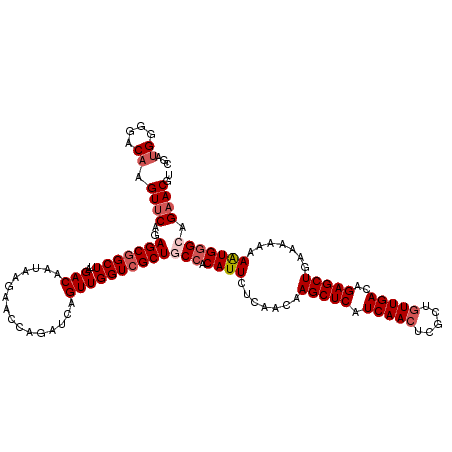
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:54 2006