
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,742,762 – 21,742,910 |
| Length | 148 |
| Max. P | 0.948739 |

| Location | 21,742,762 – 21,742,870 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -38.99 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.06 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21742762 108 - 23771897 CGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC------G------AUUGCGAGAGCGAGAGAACAGCGAAAG ......((((((..(....((((.((.......((((((.....))))))......((((((((((.............)------)------)))))))).))))))...)..)))))) ( -31.72) >DroSec_CAF1 24648 108 - 1 CGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC------G------AUUGCGAGAGCGAGAGAGCAGCGAAAG ((...((((((((((....)))).((.......((((((.....))))))......((((((((((.............)------)------)))))))).))..))))))..)).... ( -35.12) >DroSim_CAF1 29534 108 - 1 CGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC------G------AUUGCGAGAGCGAGAGAGCAGCGAAAG ((...((((((((((....)))).((.......((((((.....))))))......((((((((((.............)------)------)))))))).))..))))))..)).... ( -35.12) >DroEre_CAF1 11007 114 - 1 CGACGGCUUUCGAAGGAGACUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAGAACUUCGCGAUCGGUCGGAUAUAAUGCGGCUGCG------AUUGCGAGAGCGAGAGAGCAGCGAAAG ((...((((((((((....)))).((..((...((((((.....))))))..))..((((((((((((((.((...)).))))..))------)))))))).))..))))))..)).... ( -46.60) >DroYak_CAF1 82843 120 - 1 CGACGGCUUUCGAAGGAAACUUCAGUAAUCACGUGGCGCACAUAGUGCCGAAGAACUUCGCGAUCGGUCGGAUAUUAUGCGCUUGCGAUUCCGAUUGCGAGAGCGAGAGAGCAGCGAAAG ((...((((((((((....))))........((((((((.....))))).......((((((((((((((.(...........).)))..))))))))))).))).))))))..)).... ( -46.40) >consensus CGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC______G______AUUGCGAGAGCGAGAGAGCAGCGAAAG ((...((((((((((....)))).(((....((((((((.....)))))(((....))))))(((.....)))....)))..............((((....))))))))))..)).... (-31.26 = -31.06 + -0.20)



| Location | 21,742,790 – 21,742,910 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -34.34 |
| Energy contribution | -34.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21742790 120 + 23771897 GCAUUAUAUCCGACCGAUCGCGAAGUUUUUCGGCACUAUGUGCGCCACGUGAUUGCUGAAAUCUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCGAUAAUUCGGAUUUAAGGUGUCAUGUG (((((..((((((...(((((((((..((((((((.((((((...))))))..))))))))....))))...(((....))).........)))))...))))))....)))))...... ( -35.90) >DroSec_CAF1 24676 120 + 1 GCAUUAUAUCCGACCGAUCGCGAAGUUUUUCGGCACUAUGUGCGCCACGUGAUUGCUGAAAUCUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCGAUAAUUCGGAUUUAAGGUGUCAUGUG (((((..((((((...(((((((((..((((((((.((((((...))))))..))))))))....))))...(((....))).........)))))...))))))....)))))...... ( -35.90) >DroSim_CAF1 29562 120 + 1 GCAUUAUAUCCGACCGAUCGCGAAGUUUUUCGGCACUAUGUGCGCCACGUGAUUGCUGAAAUCUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCGAUAAUUCGGAUUUAAGGUGUCAUGUG (((((..((((((...(((((((((..((((((((.((((((...))))))..))))))))....))))...(((....))).........)))))...))))))....)))))...... ( -35.90) >DroEre_CAF1 11041 120 + 1 GCAUUAUAUCCGACCGAUCGCGAAGUUCUUCGGCACUAUGUGCGCCACGUGAUUGCUGAAGUCUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCAAUAAUUCGGAUUUAAGGUGUCAUGUG (((((..(((((((.(....(((((..((((((((.((((((...))))))..))))))))....)))))....).)))(((........)))........))))....)))))...... ( -33.50) >DroYak_CAF1 82883 120 + 1 GCAUAAUAUCCGACCGAUCGCGAAGUUCUUCGGCACUAUGUGCGCCACGUGAUUACUGAAGUUUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCGAUAAUUCGGAUUUAAAGUGUCAUGUG (((((...(((((....))).))(((.(....).))).)))))..((((((((.(((......(((..(....)..((((((........)))))).....))).....))))))))))) ( -32.00) >consensus GCAUUAUAUCCGACCGAUCGCGAAGUUUUUCGGCACUAUGUGCGCCACGUGAUUGCUGAAAUCUCCUUCGAAAGCCGUCGCUAUAUCAUCAGCGAUAAUUCGGAUUUAAGGUGUCAUGUG (((((..((((((...(((((((((..((((((((.((((((...))))))..))))))))....))))...(((....))).........)))))...))))))....)))))...... (-34.34 = -34.46 + 0.12)



| Location | 21,742,790 – 21,742,910 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -32.74 |
| Energy contribution | -31.94 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21742790 120 - 23771897 CACAUGACACCUUAAAUCCGAAUUAUCGCUGAUGAUAUAGCGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC ...........(((.((((((...(((((....((...(((....))).))((((..((((.....))))...((((((.....)))))).....)))))))))...)))))).)))... ( -32.90) >DroSec_CAF1 24676 120 - 1 CACAUGACACCUUAAAUCCGAAUUAUCGCUGAUGAUAUAGCGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC ...........(((.((((((...(((((....((...(((....))).))((((..((((.....))))...((((((.....)))))).....)))))))))...)))))).)))... ( -32.90) >DroSim_CAF1 29562 120 - 1 CACAUGACACCUUAAAUCCGAAUUAUCGCUGAUGAUAUAGCGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC ...........(((.((((((...(((((....((...(((....))).))((((..((((.....))))...((((((.....)))))).....)))))))))...)))))).)))... ( -32.90) >DroEre_CAF1 11041 120 - 1 CACAUGACACCUUAAAUCCGAAUUAUUGCUGAUGAUAUAGCGACGGCUUUCGAAGGAGACUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAGAACUUCGCGAUCGGUCGGAUAUAAUGC ...........(((.((((((...((((((...((...(((....))).))((((....))))))))))..((((((((.....)))))(((....)))))).....)))))).)))... ( -35.10) >DroYak_CAF1 82883 120 - 1 CACAUGACACUUUAAAUCCGAAUUAUCGCUGAUGAUAUAGCGACGGCUUUCGAAGGAAACUUCAGUAAUCACGUGGCGCACAUAGUGCCGAAGAACUUCGCGAUCGGUCGGAUAUUAUGC ...............((((((...(((((((((((...(((....))).))((((....))))....))))...(((((.....)))))(((....))))))))...))))))....... ( -36.40) >consensus CACAUGACACCUUAAAUCCGAAUUAUCGCUGAUGAUAUAGCGACGGCUUUCGAAGGAGAUUUCAGCAAUCACGUGGCGCACAUAGUGCCGAAAAACUUCGCGAUCGGUCGGAUAUAAUGC ...............((((((...(((((....((...(((....))).))((((....((((.((......))(((((.....)))))))))..)))))))))...))))))....... (-32.74 = -31.94 + -0.80)

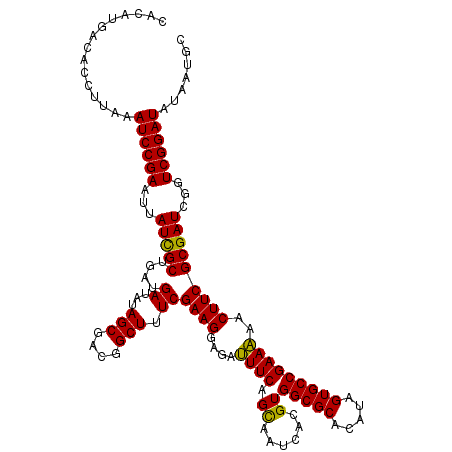
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:49 2006