
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,742,388 – 21,742,545 |
| Length | 157 |
| Max. P | 0.999456 |

| Location | 21,742,388 – 21,742,505 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -32.94 |
| Energy contribution | -32.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
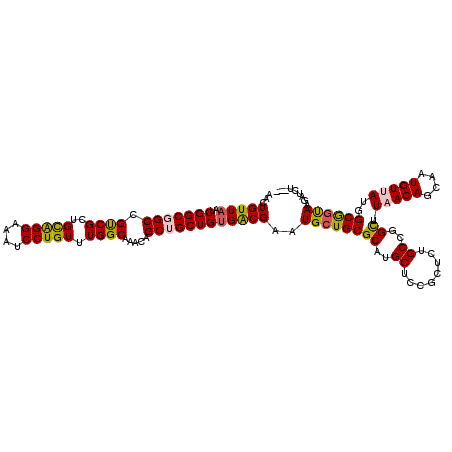
>3L_DroMel_CAF1 21742388 117 - 23771897 AACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGGCGAAUGGUGCGCGUGCUCCGUUCUGCCGGUUUAACAGCAAUGUUAUGGCGGUAAGAUCU--- .(((((((((((((((((((((..(((((....))))).))))..((((...)))))))((((((.((....)).))))))))))))).(((((....)))))))))))).......--- ( -47.00) >DroSec_CAF1 24257 117 - 1 AACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCUCAGCAAGCUUAACAGCAAUGUUAUGGCGGUAAGAUCU--- ........(((....((((((((((((((....)))))..))).((((..((((((...(..(((((.((.......)).)))))..)...)))))).))))..))))))..)))..--- ( -43.30) >DroSim_CAF1 29142 117 - 1 AACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCUCUGCAGGCUUAACAGCAAUGUUAUGGCGGUAAGAUCU--- ...(((((..(((((((.((.((((((((....)))))..)))..)).))))))))))))..((((((((.(((........))).)).(((((....)))))..))))))......--- ( -41.30) >DroEre_CAF1 10618 117 - 1 AACUGUUAAUCGGCCGCCGCCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCUGCUCUGCCGGCUUUACAGAAAUGUUAAUGCGGUAAGAUCU--- ........(((.(((((.((((..(((((....))))).)))).((((....((((((.....((((.(((.((...))..))))))).))))))...))))...)))))..)))..--- ( -38.80) >DroYak_CAF1 82448 120 - 1 AACUGUUAAUCGGCCGCCGCCGUUGUGGGAAAUCCUGUUUGGCAAACAGCUGCUGCGACGAAUGCUGCGCAUGCUCCGCUCUGCCGGUUUUACAGCAAUGUUAAGGCAGCAAGAUCUGCA ....((..(((.((.(((((((..(..((....))..).)))).((((..(((((..(.....((((.(((.((...))..))))))).)..))))).))))..))).))..)))..)). ( -37.50) >consensus AACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCUCUGCCGGCUUAACAGCAAUGUUAUGGCGGUAAGAUCU___ ...(((((..(((((((.((((..(((((....))))).)))).....))))))))))))..((((((((..((........))..)).(((((....)))))..))))))......... (-32.94 = -32.86 + -0.08)


| Location | 21,742,425 – 21,742,545 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -50.84 |
| Consensus MFE | -46.06 |
| Energy contribution | -46.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21742425 120 - 23771897 GUGCGUGCGUUUGCAACAGUGGCAGGGCACGGCGAAACUUAACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGGCGAAUGGUGCGCGUGCUCCGUU (..(((((((((((.((((..((..(((.((.((((((......)))..))).)))))((((..(((((....))))).)))).....))..)))).))))))...)))))..)...... ( -55.10) >DroSec_CAF1 24294 120 - 1 GUGUGUGCGUUUGCAACAGUGGCAGGGCACGGCGAAACUUAACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCU (((((((((((((..((((..((..(((.((.((((((......)))..))).)))))((((..(((((....))))).)))).....))..))))..))))))).))))))........ ( -52.40) >DroSim_CAF1 29179 120 - 1 GUGUGUGCGUUUGCAACAGUGGCAGGGCACGGCGAAACUUAACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCU (((((((((((((..((((..((..(((.((.((((((......)))..))).)))))((((..(((((....))))).)))).....))..))))..))))))).))))))........ ( -52.40) >DroEre_CAF1 10655 120 - 1 GUGUGUGCGUUUGCAACAGUGGCAGGGCACGGCGAAACUUAACUGUUAAUCGGCCGCCGCCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCUGCU (((((((((((((..((((..((..(((..((((((((......)))..)).))))))((((..(((((....))))).)))).....))..))))..))))))).))))))........ ( -51.20) >DroYak_CAF1 82488 120 - 1 GUGUGUGCGUUUGCAACAGUGGCAGGACACGGCGAAACUUAACUGUUAAUCGGCCGCCGCCGUUGUGGGAAAUCCUGUUUGGCAAACAGCUGCUGCGACGAAUGCUGCGCAUGCUCCGCU (((((((((((((...(((..((.((...(((((((((......)))..)).))))))((((..(..((....))..).)))).....))..)))...))))))).))))))........ ( -43.10) >consensus GUGUGUGCGUUUGCAACAGUGGCAGGGCACGGCGAAACUUAACUGUUAAUCGGCGGCCGUCGCUGCAGGAAAUCCUGUUUGGCAAACAGCUGCUGUGACGAAUGCUGCGCAUGCUCCGCU (((((((((((((..((((..((..(((.((.((((((......)))..))).)))))((((..(((((....))))).)))).....))..))))..))))))).))))))........ (-46.06 = -46.18 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:46 2006