| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,740,849 – 21,740,968 |
| Length | 119 |
| Max. P | 0.995044 |

| Location | 21,740,849 – 21,740,968 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -35.18 |
| Energy contribution | -35.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740849 119 + 23771897 CAGCAGCUCCAGGAGCUGCAUCGCCACUUGGCAGAUUAUGACGACAACUAUACUGACAGGCAGAGCCAGAGGAAACUACUCCAAGAAACUAGCCAGAUUCAGUUGCAGUCGCAAUCGGA- ..(((((((...)))))))...(((....))).((((.((((..(((((...(((.(.(((...)))...(((......))).........).)))....)))))..)))).))))...- ( -38.20) >DroSec_CAF1 22752 120 + 1 CAGCAGCUCCAGGAGCUGCAUCGCCACUUGGCAGAUUAUGACGACAACUAUGCUGACAGGCAGAGCCAGAGGGAACUACUCCAAGAAACUAGCCAGAUUCAGUUGCAGUCGCAGUCGGAG ..(((((((...))))))).(((((....))).((((.((((..(((((...(((.(.(((...)))....(((.....))).........).)))....)))))..)))).)))).)). ( -39.00) >DroSim_CAF1 27629 120 + 1 CAGCAGCUCCAGGAGCUGCAUCGCCACUUGGCAGAUUAUGACGACAACUAUGCUGACAGGCAGAGCCAGAGGAAACUACUCCAAGAAACUAGCCAGAUUCAGUUGCAGUCGCAGUCGGAG ..(((((((...))))))).(((((....))).((((.((((..(((((...(((.(.(((...)))...(((......))).........).)))....)))))..)))).)))).)). ( -39.40) >DroEre_CAF1 8877 120 + 1 CAGCAGCUCCAGGAGCUGCAUCGCCAGUUGGCAGAUUAUGACGACAACUAUGCUGACAGGCAGCGCCAGAAGAAACUACUCCAAGAAACUAGCCAGAUUUUGUGGCAGUUGCAGUCGGAU ..(((((((...)))))))((((((....)))......((((..(((((..((((.....))))((((.((((..((......))...((....)).)))).)))))))))..))))))) ( -39.40) >DroYak_CAF1 80788 120 + 1 CAGCAGCUUCAGGAGCUGCAUCGCCAGUUGGCAGAUUAUGACGACAACUAUGCUGACAGGCAGAGCCAGAGGAAACUACUCCAAGAAACUAGCCAGAUUUUGUUGCAGUUGCAGUCGGAU ..(((((((...)))))))((((((....))).((((..(((..((((....(((.(.(((...)))...(((......))).........).))).....))))..)))..)))).))) ( -35.20) >consensus CAGCAGCUCCAGGAGCUGCAUCGCCACUUGGCAGAUUAUGACGACAACUAUGCUGACAGGCAGAGCCAGAGGAAACUACUCCAAGAAACUAGCCAGAUUCAGUUGCAGUCGCAGUCGGAG ..(((((((...)))))))...(((....))).((((..(((..(((((...(((.(.(((...)))...(((......))).........).)))....)))))..)))..)))).... (-35.18 = -35.26 + 0.08)

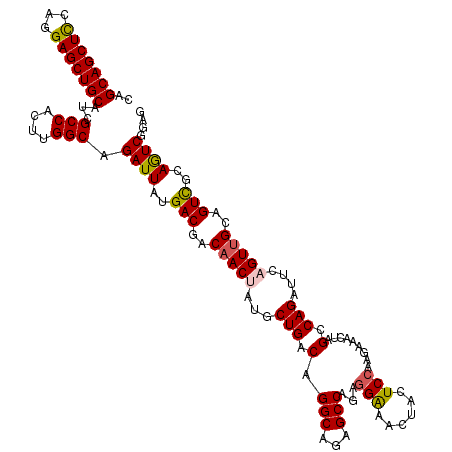

| Location | 21,740,849 – 21,740,968 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -38.12 |
| Energy contribution | -38.76 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740849 119 - 23771897 -UCCGAUUGCGACUGCAACUGAAUCUGGCUAGUUUCUUGGAGUAGUUUCCUCUGGCUCUGCCUGUCAGUAUAGUUGUCGUCAUAAUCUGCCAAGUGGCGAUGCAGCUCCUGGAGCUGCUG -...(((.(((((((..(((((....(((.((((....((((....))))...))))..)))..))))).))))))).)))...(((.(((....))))))(((((((...))))))).. ( -43.90) >DroSec_CAF1 22752 120 - 1 CUCCGACUGCGACUGCAACUGAAUCUGGCUAGUUUCUUGGAGUAGUUCCCUCUGGCUCUGCCUGUCAGCAUAGUUGUCGUCAUAAUCUGCCAAGUGGCGAUGCAGCUCCUGGAGCUGCUG ....(((.(((((((...((((....(((.((((....((((...))))....))))..)))..))))..))))))).)))...(((.(((....))))))(((((((...))))))).. ( -41.70) >DroSim_CAF1 27629 120 - 1 CUCCGACUGCGACUGCAACUGAAUCUGGCUAGUUUCUUGGAGUAGUUUCCUCUGGCUCUGCCUGUCAGCAUAGUUGUCGUCAUAAUCUGCCAAGUGGCGAUGCAGCUCCUGGAGCUGCUG ....(((.(((((((...((((....(((.((((....((((....))))...))))..)))..))))..))))))).)))...(((.(((....))))))(((((((...))))))).. ( -43.70) >DroEre_CAF1 8877 120 - 1 AUCCGACUGCAACUGCCACAAAAUCUGGCUAGUUUCUUGGAGUAGUUUCUUCUGGCGCUGCCUGUCAGCAUAGUUGUCGUCAUAAUCUGCCAACUGGCGAUGCAGCUCCUGGAGCUGCUG .(((((..(.((((((((.......)))).)))).))))))((((((((....((.(((((..(((.((.((((((.((........)).)))))))))))))))).)).)))))))).. ( -41.30) >DroYak_CAF1 80788 120 - 1 AUCCGACUGCAACUGCAACAAAAUCUGGCUAGUUUCUUGGAGUAGUUUCCUCUGGCUCUGCCUGUCAGCAUAGUUGUCGUCAUAAUCUGCCAACUGGCGAUGCAGCUCCUGAAGCUGCUG ....(((.(((((((...........((((((......((((....)))).)))))).(((......)))))))))).)))...(((.(((....))))))((((((.....)))))).. ( -36.70) >consensus AUCCGACUGCGACUGCAACUGAAUCUGGCUAGUUUCUUGGAGUAGUUUCCUCUGGCUCUGCCUGUCAGCAUAGUUGUCGUCAUAAUCUGCCAAGUGGCGAUGCAGCUCCUGGAGCUGCUG ....(((.(((((((...((((....(((.((..((..((((....))))...))..)))))..))))..))))))).)))...(((.(((....))))))(((((((...))))))).. (-38.12 = -38.76 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:42 2006