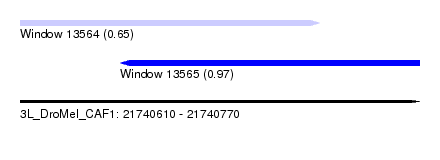
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,740,610 – 21,740,770 |
| Length | 160 |
| Max. P | 0.965796 |

| Location | 21,740,610 – 21,740,730 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -26.50 |
| Energy contribution | -27.18 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740610 120 + 23771897 UCCGUAUCGCUUCAAGUGGCCACAAUUUGAGGCAGAUUGCGCUCUAAUAAAUAUGGGCAGCCAUCUAAGCUAAUGCCCCAACUCAUCACCGAGUUGAGAGAUGAGCACGCAAGCAGGCGA ..(((...(((((((((.......))))))))).(.((((((((..........)))).(((((((..((....))..((((((......))))))..))))).))..)))).)..))). ( -38.90) >DroSec_CAF1 22513 120 + 1 UUCGUAUCGCUUCAAGUGGCCACAAUUUGAGGCAGAUUGCGCUCUAAUAAAUAUGGGCAGCCAUCAAAGCUAAUGCCCCAACUCAUCACCGAGUUGGGAGAUGAGCACGCGAGCAGGCAA ...((...(((((((((.......))))))))).(.((((((((..........)))).((((((...((....))((((((((......)))))))).)))).))..)))).)..)).. ( -42.60) >DroSim_CAF1 27390 120 + 1 UUCGUAUCGCUCCAAGUGGCCACAAUUUGAGGCAGAUUGCGCUCUAAUAAAUAUGGGCAGCCAUCAAAGCUAAUGCCCCAACUCAUCACCGAGUUGGGAGAUGAGCACGCGAGCAGGCGA .((((...((((......(((.(.....).))).....((((((..........)))).((((((...((....))((((((((......)))))))).)))).))..))))))..)))) ( -43.00) >DroEre_CAF1 8642 115 + 1 UCUAUAUCGCUCCAAGUGGCCACAAUUUGAGGCAGAUAGCGCUCUAAUAAAUAUGUGCAGCCAUCAAAGCUAAUGCCCCAACUCAUCACCG-----GGAGAUGAACACGCGAGCAGGCCA ........((((...(((........(((.((((....((((............))))(((.......)))..)))).))).(((((....-----...))))).)))..))))...... ( -27.50) >DroYak_CAF1 80553 115 + 1 UCUGUAUCGCUUCCAGUGGCAAAAAUUUGAGGCAGAUUGCGCUCUAAUAAAUAUGAGCAACCAUCAAAGCUAAUGCACCAACUCAUCACCG-----GGAGAUGAACACGAGAGCAGGCCA (((((.((((((((.((((......(((((.((.....))((((..........)))).....)))))((....)).........)))).)-----)))).......)))..)))))... ( -26.51) >consensus UCCGUAUCGCUUCAAGUGGCCACAAUUUGAGGCAGAUUGCGCUCUAAUAAAUAUGGGCAGCCAUCAAAGCUAAUGCCCCAACUCAUCACCGAGUUGGGAGAUGAGCACGCGAGCAGGCCA ........((((...(((........(((.(((((((.((((((..........)))).)).)))........)))).)))((((((.((......)).)))))))))..))))...... (-26.50 = -27.18 + 0.68)

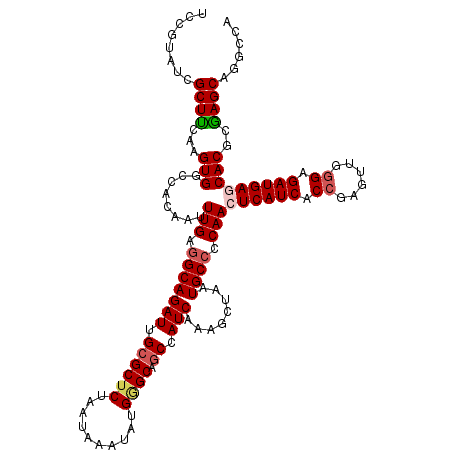
| Location | 21,740,650 – 21,740,770 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -35.30 |
| Energy contribution | -35.62 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740650 120 - 23771897 UUAAUUAAGUUUGGGCCUUGGCAACGGGCGCAUGCGUAUCUCGCCUGCUUGCGUGCUCAUCUCUCAACUCGGUGAUGAGUUGGGGCAUUAGCUUAGAUGGCUGCCCAUAUUUAUUAGAGC (((((.((((.(((((....((((((((((...........)))))).))))..((.((((((((((((((....)))))))))((....))..))))))).))))).)))))))))... ( -43.30) >DroSec_CAF1 22553 120 - 1 UUAAUUAAGUUUGGGCCUUGGCAACGGGCGCAUGCGUAUCUUGCCUGCUCGCGUGCUCAUCUCCCAACUCGGUGAUGAGUUGGGGCAUUAGCUUUGAUGGCUGCCCAUAUUUAUUAGAGC (((((.((((.(((((..((....))((((((((((...(......)..))))))).((((.(((((((((....)))))))))((....))...)))))))))))).)))))))))... ( -44.90) >DroSim_CAF1 27430 120 - 1 UUAAUUAAGUUUGGGCCUUGGCAACGGGCGCAUGCGUAUCUCGCCUGCUCGCGUGCUCAUCUCCCAACUCGGUGAUGAGUUGGGGCAUUAGCUUUGAUGGCUGCCCAUAUUUAUUAGAGC (((((.((((.(((((...(((.(((.(.(((.(((.....))).))).).)))(((.((.((((((((((....)))))))))).)).))).......)))))))).)))))))))... ( -45.90) >DroEre_CAF1 8682 115 - 1 UUAAUUAAGUUUGGGCCUUGGCAACGGGCGCAUGCGUAUCUGGCCUGCUCGCGUGUUCAUCUCC-----CGGUGAUGAGUUGGGGCAUUAGCUUUGAUGGCUGCACAUAUUUAUUAGAGC (((((.((((.((.((((..((.(((.(.(((.((.......)).))).).)))..(((((...-----....)))))))..))))..(((((.....)))))..)).)))))))))... ( -34.50) >DroYak_CAF1 80593 115 - 1 UUAAUUAAGUUUGGGCCUUGGCAACGGGCUCAUGCGUAUCUGGCCUGCUCUCGUGUUCAUCUCC-----CGGUGAUGAGUUGGUGCAUUAGCUUUGAUGGUUGCUCAUAUUUAUUAGAGC ........((.(((((((.(....)))))))).)).......(((.((((......(((((...-----.))))).)))).)))......((((((((((..........)))))))))) ( -32.70) >consensus UUAAUUAAGUUUGGGCCUUGGCAACGGGCGCAUGCGUAUCUCGCCUGCUCGCGUGCUCAUCUCCCAACUCGGUGAUGAGUUGGGGCAUUAGCUUUGAUGGCUGCCCAUAUUUAUUAGAGC (((((.((((.(((((((..((.(((.(.(((.(((.....))).))).).)))..(((((.((......)).)))))))..))))..(((((.....))))).))).)))))))))... (-35.30 = -35.62 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:40 2006