
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,740,056 – 21,740,251 |
| Length | 195 |
| Max. P | 0.999931 |

| Location | 21,740,056 – 21,740,176 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -34.36 |
| Energy contribution | -36.20 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740056 120 + 23771897 GGCAUUCCGGCCUCAAGUCAGCUUUGGAAGAUUUGUCGCCUAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG ........((((..((..(((((.(((((((...(((((......((((.......))))))))).))))))).))......)))..))...))))((((((((.......)))))))). ( -38.00) >DroSec_CAF1 21951 120 + 1 GGCAUUCCGGCUUCAAGUCAGCUUCGGAAGAUUUGUCGCCUAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG (((...(((((..((((((..(....)..))))))..))).......((((.....))))((((......))))..........)).......)))((((((((.......)))))))). ( -39.80) >DroSim_CAF1 26838 120 + 1 GGCAUUCCGGCUUCAAGUCAGCUUCGGAAGAUUUGUCGCCUAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG (((...(((((..((((((..(....)..))))))..))).......((((.....))))((((......))))..........)).......)))((((((((.......)))))))). ( -39.80) >DroEre_CAF1 7922 120 + 1 GGCAUUCCGGCUUCAAGUCAGCUUCGGAAGAUUUGUCGCCUAUUUUGUUUGUGCCUCGCAGUGAUUUCUCUCGUAGAUUCGUCUUGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG ((((....(((..((((((..(....)..))))))..))).....))))...(((..((((.(((.(((.....)))...))))))).....)))(((((((((.......))))))))) ( -34.70) >DroYak_CAF1 79587 120 + 1 GGUAUUCCGGCUUCAAGUCAGCUUCGGAAGAUUUGUCGCCUAUUUUGUUUGUGCCUCGCAGUGAUUUCUUUCGCAGAUUCGUCUUCUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG ((....))(((..((((((..(....)..))))))..)))............(((.....((((......))))(((.......))).....)))(((((((((.......))))))))) ( -36.30) >consensus GGCAUUCCGGCUUCAAGUCAGCUUCGGAAGAUUUGUCGCCUAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUG (((((...(((..((((((..(....)..))))))..)))..........)))))..((.((((......))))(((....))).........))(((((((((.......))))))))) (-34.36 = -36.20 + 1.84)

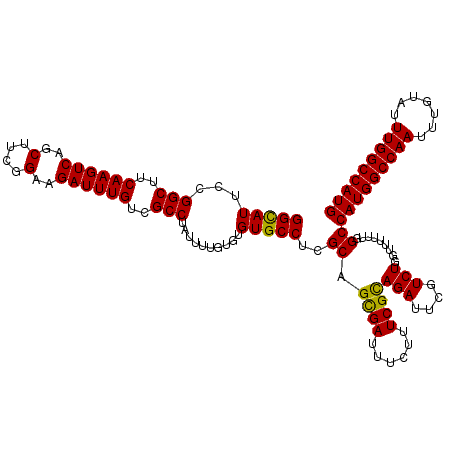

| Location | 21,740,056 – 21,740,176 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.19 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740056 120 - 23771897 CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUAGGCGACAAAUCUUCCAAAGCUGACUUGAGGCCGGAAUGCC (((((((((.......)))))))))(((........(((....)))((((......)))).....)))............((((.......((((...(((.......))).)))))))) ( -30.51) >DroSec_CAF1 21951 120 - 1 CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUAGGCGACAAAUCUUCCGAAGCUGACUUGAAGCCGGAAUGCC (((((((((.......)))))))))(((........(((....)))((((......)))).....)))............((((.......(((((..(((.......)))))))))))) ( -33.41) >DroSim_CAF1 26838 120 - 1 CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUAGGCGACAAAUCUUCCGAAGCUGACUUGAAGCCGGAAUGCC (((((((((.......)))))))))(((........(((....)))((((......)))).....)))............((((.......(((((..(((.......)))))))))))) ( -33.41) >DroEre_CAF1 7922 120 - 1 CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACAAGACGAAUCUACGAGAGAAAUCACUGCGAGGCACAAACAAAAUAGGCGACAAAUCUUCCGAAGCUGACUUGAAGCCGGAAUGCC (((((((((.......)))))))))(((...........((......)).((....)).......)))............((((.......(((((..(((.......)))))))))))) ( -31.11) >DroYak_CAF1 79587 120 - 1 CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAAGAAGACGAAUCUGCGAAAGAAAUCACUGCGAGGCACAAACAAAAUAGGCGACAAAUCUUCCGAAGCUGACUUGAAGCCGGAAUACC (((((((((.......)))))))))(((........(((....))).(....)............)))............(((..(((.((..........)).)))..)))........ ( -28.10) >consensus CAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUAGGCGACAAAUCUUCCGAAGCUGACUUGAAGCCGGAAUGCC (((((((((.......)))))))))(((.......((((....))))(....)............)))............((((.......(((((..(((.......)))))))))))) (-28.55 = -29.19 + 0.64)

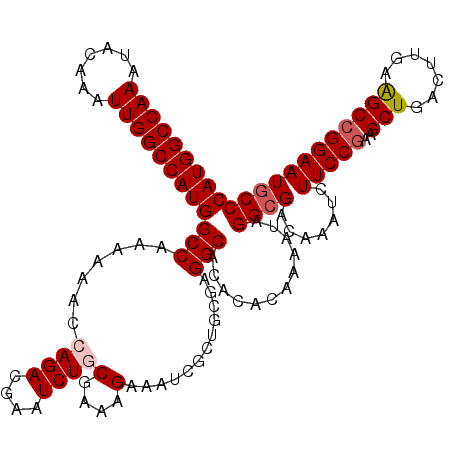

| Location | 21,740,096 – 21,740,216 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740096 120 + 23771897 UAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCAAGUGUUUGCCAAUCU ..(((((((((((((.....((((......))))(((....))))))....((((.((((((((.......))))))))))))))))))))))(((((((.....)).)))))....... ( -43.20) >DroSec_CAF1 21991 120 + 1 UAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCGAGUGUUUGCCAAUCU ..(((((((((((((.....((((......))))(((....))))))....((((.((((((((.......))))))))))))))))))))))(((((((.....)).)))))....... ( -42.50) >DroSim_CAF1 26878 120 + 1 UAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCGAGUGUUUGCCAAUCU ..(((((((((((((.....((((......))))(((....))))))....((((.((((((((.......))))))))))))))))))))))(((((((.....)).)))))....... ( -42.50) >DroEre_CAF1 7962 120 + 1 UAUUUUGUUUGUGCCUCGCAGUGAUUUCUCUCGUAGAUUCGUCUUGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUUAAAUGUUUGGCAAUCU ...........((((..((((.((....)).).(((((..((((.((((((((((.((((((((.......))))))))))))).....)))))))))..)))))..)))..)))).... ( -35.70) >DroYak_CAF1 79627 120 + 1 UAUUUUGUUUGUGCCUCGCAGUGAUUUCUUUCGCAGAUUCGUCUUCUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUUAAGUGUUUGGCAAUCU ....((((((((((...((.((((......)))).................((((.((((((((.......))))))))))))))))))))..(((((((.....)).)))))))))... ( -38.20) >consensus UAUUUUGUGUGUGCCUCGCAGCGAUUUCUUUCGCAGAUUCGUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCAAGUGUUUGCCAAUCU ..((((((((((........((((......)))).................((((.((((((((.......))))))))))))))))))))))(((((((.....)).)))))....... (-36.00 = -36.20 + 0.20)



| Location | 21,740,096 – 21,740,216 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -30.22 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740096 120 - 23771897 AGAUUGGCAAACACUUGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUA (((((..(((....)))....)))))((((((((.((((((((((((((.......)))))))).)))......((((.(((.....(....)...)))))).).)))...)))))))). ( -36.30) >DroSec_CAF1 21991 120 - 1 AGAUUGGCAAACACUCGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUA ...(((.......((((...((((.(((((.((....((((((((((((.......)))))))).))))....)).))))).))))((((......))))..)))).......))).... ( -36.34) >DroSim_CAF1 26878 120 - 1 AGAUUGGCAAACACUCGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUA ...(((.......((((...((((.(((((.((....((((((((((((.......)))))))).))))....)).))))).))))((((......))))..)))).......))).... ( -36.34) >DroEre_CAF1 7962 120 - 1 AGAUUGCCAAACAUUUAAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACAAGACGAAUCUACGAGAGAAAUCACUGCGAGGCACAAACAAAAUA ....((((............((((.((((((((....((((((((((((.......)))))))).))))....)))))))).))))....((....)).......))))........... ( -37.70) >DroYak_CAF1 79627 120 - 1 AGAUUGCCAAACACUUAAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAAGAAGACGAAUCUGCGAAAGAAAUCACUGCGAGGCACAAACAAAAUA ....((((...........(((((.((((((.(....((((((((((((.......)))))))).))))....).)))))).)))))(....)............))))........... ( -35.40) >consensus AGAUUGGCAAACACUUGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGACGAAUCUGCGAAAGAAAUCGCUGCGAGGCACACACAAAAUA ...(((.......((((..(((((.(((((.((....((((((((((((.......)))))))).))))....)).))))).)))))(....).........)))).......))).... (-30.22 = -30.62 + 0.40)

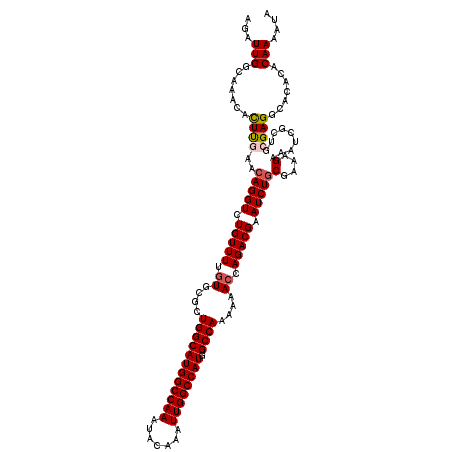

| Location | 21,740,136 – 21,740,251 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -30.22 |
| Energy contribution | -31.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740136 115 + 23771897 GUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCAAGUGUUUGCCAAUCUCCGAUUGGCUUGACUGAAGUUUUCACUGAAAAUUU----- (((((.(((((((((.((((((((.......))))))))))))).....)))))))))...((((.(((...(((((((...)))))))..(((....)))..))))))).....----- ( -37.50) >DroSec_CAF1 22031 115 + 1 GUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCGAGUGUUUGCCAAUCUCCGAUUGGCUUGACUGGAGUUUUCUCUGAAAAUUU----- (((((.(((((((((.((((((((.......))))))))))))).....)))))))))...(((....(((.(((((((...)))))))..))).((((....))))))).....----- ( -38.80) >DroSim_CAF1 26918 115 + 1 GUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCGAGUGUUUGCCAAUCUCCGAUUGGCUUGACUGGAGUUUUCUCUGAAAAUUU----- (((((.(((((((((.((((((((.......))))))))))))).....)))))))))...(((....(((.(((((((...)))))))..))).((((....))))))).....----- ( -38.80) >DroEre_CAF1 8002 120 + 1 GUCUUGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUUAAAUGUUUGGCAAUCUCCGAUUGGCUUGGCUGAAGUUUUCUCUGAAAAGUUUUAGG (((..(((..((((..((((((((.......))))))))(((((.(((..(((((....)))))...)))))))).....))))..)))..))).(((.(((((...))))).))).... ( -33.80) >DroYak_CAF1 79667 115 + 1 GUCUUCUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUUAAGUGUUUGGCAAUCUCCGAUUG-----ACUGAAGUUUUCUUUAAAAAGUUUGAGG (((.......(((((.((((((((.......))))))))))))).((((.(((((....)))))..))))..)))...(((.((((.-----..(((((....)))))...)))).))). ( -33.80) >consensus GUCUGGUUUUUUGGCCAUGGCCAAUUUGUAUUUGGCCAUGCCAGCGCACAAAACAGACCUGUUCAAGUGUUUGCCAAUCUCCGAUUGGCUUGACUGAAGUUUUCUCUGAAAAUUU_____ ((((.((((((((((.((((((((.......))))))))))))).....)))))))))..........(((.(((((((...)))))))..)))..((((((((...))))))))..... (-30.22 = -31.42 + 1.20)



| Location | 21,740,136 – 21,740,251 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -30.22 |
| Energy contribution | -30.02 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21740136 115 - 23771897 -----AAAUUUUCAGUGAAAACUUCAGUCAAGCCAAUCGGAGAUUGGCAAACACUUGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGAC -----........(((....)))...(((..(((((((...)))))))...(((..(((((....))))).)))...((((((((((((.......)))))))).))))........))) ( -36.00) >DroSec_CAF1 22031 115 - 1 -----AAAUUUUCAGAGAAAACUCCAGUCAAGCCAAUCGGAGAUUGGCAAACACUCGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGAC -----.........(((....)))..(((..(((((((...)))))))...(((..(((((....))))).)))...((((((((((((.......)))))))).))))........))) ( -35.00) >DroSim_CAF1 26918 115 - 1 -----AAAUUUUCAGAGAAAACUCCAGUCAAGCCAAUCGGAGAUUGGCAAACACUCGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGAC -----.........(((....)))..(((..(((((((...)))))))...(((..(((((....))))).)))...((((((((((((.......)))))))).))))........))) ( -35.00) >DroEre_CAF1 8002 120 - 1 CCUAAAACUUUUCAGAGAAAACUUCAGCCAAGCCAAUCGGAGAUUGCCAAACAUUUAAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACAAGAC (((....(((....))).........(.(((.((....))...))).)............)))...(((((((....((((((((((((.......)))))))).))))....))))))) ( -28.80) >DroYak_CAF1 79667 115 - 1 CCUCAAACUUUUUAAAGAAAACUUCAGU-----CAAUCGGAGAUUGCCAAACACUUAAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAAGAAGAC .....................((((.(.-----(((((...))))).)...(((..(((((....))))).)))...((((((((((((.......)))))))).)))).....)))).. ( -29.20) >consensus _____AAAUUUUCAGAGAAAACUUCAGUCAAGCCAAUCGGAGAUUGGCAAACACUUGAACAGGUCUGUUUUGUGCGCUGGCAUGGCCAAAUACAAAUUGGCCAUGGCCAAAAAACCAGAC .......................(((((...(((((((...)))))))...(((..(((((....))))).))).)))))(((((((((.......)))))))))............... (-30.22 = -30.02 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:38 2006