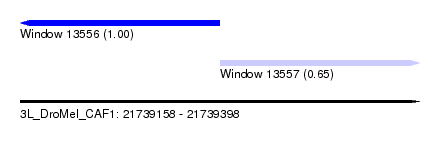
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,739,158 – 21,739,398 |
| Length | 240 |
| Max. P | 0.999272 |

| Location | 21,739,158 – 21,739,278 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21739158 120 - 23771897 AAAACAUCAUCUUGAACCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAACAGGACUCGUUCAUCUGCCACGGAACUUCAGCCAAGCAGCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.((((.....))))..........((((...((........))..)))).)))).)))))))))))..))))). ( -28.30) >DroSec_CAF1 21053 120 - 1 AAAACAUCAUCUUGAACCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAACAGGACUCUUUCAUCUGCCACGGAACUCCAGCCAAGCAGCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.((((.....))))..........((((...((........))..)))).)))).)))))))))))..))))). ( -28.30) >DroSim_CAF1 25945 120 - 1 AAAACAUCAUCUUGAACCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAACAGGACUCUUUCAUCUGCCACGGAACUCCAGCCAAGCAGCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.((((.....))))..........((((...((........))..)))).)))).)))))))))))..))))). ( -28.30) >DroEre_CAF1 7129 120 - 1 AAAACAUCAUCUUGAACCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAGCAGGACUCUUUCAUCUGCCAUGGAACUCCAGCCAAGCAUCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.(((((...)))))...........(((..(((....))).....)))..)))).)))))))))))..))))). ( -30.00) >DroYak_CAF1 78694 120 - 1 AAAACAUCAUCUUGAAUCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAAUAGGACUCCUUCAUCUGCCAUGGAACUCCAGCCAAGCAUCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.((((.....))))...........(((..(((....))).....)))..)))).)))))))))))..))))). ( -25.70) >consensus AAAACAUCAUCUUGAACCAUUAGCAAUGUCAGACACAUUAAGCAUCCCCUGCAAAACAGGACUCUUUCAUCUGCCACGGAACUCCAGCCAAGCAGCGGUGAUUAAUGUGUUUAUUAUGCA ......................(((.((..(((((((((((.((((.((((.....))))..........((((...((........))..)))).)))).)))))))))))..))))). (-26.66 = -26.90 + 0.24)


| Location | 21,739,278 – 21,739,398 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -16.06 |
| Energy contribution | -17.34 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21739278 120 + 23771897 GAUGCAUUUCGUUUAAGUUCCCCCUCCCCAAUUUCACAAAAAAAAGCCCAAAAGUCUGAGCAUCUGCCUUUUUCUCAAUUCCUUGAGAAGCAGGGAAUGCGAACGAUUGUUGGUAUCUAA (((((...((((((..((((((.......................((.((......)).))...(((....(((((((....))))))))))))))))..))))))......)))))... ( -30.10) >DroSec_CAF1 21173 120 + 1 GAUGCAUUUCGUUUAUGUUCCCCUUCCCCCACUUCACAAAAAAAAGCCCAAAAAUCUAAGCAUCUGCCUUUUUCUCAAUCCCUUGAGAAGCAUGGAAUACGAACGAUUGUUGGCAUUUAA (((((...(((((..((((((......................................((....))...((((((((....))))))))...))))))..)))))......)))))... ( -25.00) >DroSim_CAF1 26065 120 + 1 GAUGCAUUUCGUUUAUGUUCCCCUUCCCCCACUUCACACAAAAAAGCCCACAAAUCUAAGCAUCUGCCUUUUUCUCAAUCCCUUGAGAAUCAUGGAAUACCAACGAUUGUUGGCAUUCAA (((((...(((((..((((((......................................((....))....(((((((....)))))))....))))))..)))))......)))))... ( -21.70) >DroEre_CAF1 7249 116 + 1 GAUGCAUUUCGUUUAUGUACCCCU--CCCCACUACACAAA-AAUAGCCCAAAAAUCUACGCAUCUGUCAUUUC-CCAUUUCGUUGGGAAGCAUGGAAUAUAAACGAUUUUUGGUAUCUAA (((((...((((((((((.((...--..............-....((............))........((((-(((......)))))))...)).))))))))))......)))))... ( -25.50) >DroYak_CAF1 78814 117 + 1 GAUGCAUUUCAUUUAUGUACCCCU--CCCUACUACACAAAAAAUUGCCCAAAAAUCUAAGCAUCUGCCAUUAU-CCAUUUCAUUGAGAAGCAUGGAAUAUGAACGAUUGCUGGUAUAUCA ((((((.....(((.((((.....--......)))).)))....)))(((..((((...((....))(((..(-(((((((.....)))..)))))..)))...))))..)))...))). ( -15.60) >consensus GAUGCAUUUCGUUUAUGUUCCCCUUCCCCCACUUCACAAAAAAAAGCCCAAAAAUCUAAGCAUCUGCCUUUUUCUCAAUUCCUUGAGAAGCAUGGAAUACGAACGAUUGUUGGUAUCUAA (((((...((((((.((((((......................................((....))....((((((......))))))....)))))).))))))......)))))... (-16.06 = -17.34 + 1.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:32 2006