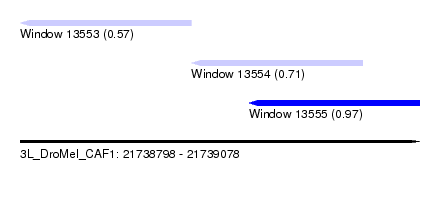
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,738,798 – 21,739,078 |
| Length | 280 |
| Max. P | 0.968848 |

| Location | 21,738,798 – 21,738,918 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -28.86 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21738798 120 - 23771897 GACAGGCCUCGUCUCACCUCAAGACAUUCACAGCCCGGAUAAAUCACACAGCGAUGAAUUUGCAUAACUCCCGGUGCAGAUCGGUCGAGUAAUUUGUUCUGCUUAAGAGGAAACUAGUUU (((((.((((((((.......))))......(((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))...))))...)).))). ( -32.20) >DroSec_CAF1 20694 120 - 1 GACAGGCCUCGUCUCACCUCAAGACAUUCCCAGCCCGGAUAAAUCACACAGCGAUGAAUUUGCAUAACUCUCGGUGCAGAUCGGUCGAGUAAUUUGUUCUGCUUAAGAGGAAACUAGUUU (((((.((((((((.......))))......(((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))...))))...)).))). ( -32.20) >DroSim_CAF1 25586 120 - 1 GACAGGCCUCGUCUCACCUCAAGACAUUCCCAGCCCGGAUAAAUCACACAGCGAUGAAUUUGCAUAACUCCCGGUGCAGAUCGGUCGAGUAAUUUGUUCUGCUUAAGAGGAAACUAGUUU (((((.((((((((.......))))......(((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))...))))...)).))). ( -32.20) >DroEre_CAF1 6773 116 - 1 GACAGGCCUCGUCUC---CCAAGACAUUCCCAGCCCGAAUAAAUCACACAGCGAUGAAUUUGCAUAACUCCUGGUGCAGAUCGGUCGAGUAAUUUGUUCUGCUUAGC-AGAAACUAGUUU (((.((..((((((.---...))))......(((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))....-.))..)).))). ( -27.00) >DroYak_CAF1 78347 116 - 1 GACAGGCUUCGUCUC---CCAAGACAUUCCCAGCCCGAAUAAAUCACACAGCGAUGAAUUUGCAUAACUCCCGGUGCAGAUCAGUCGAGUAAUUUGUUCUGCUUAAG-GGAAACUAGUUU (((((.....((((.---...)))).((((((((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))...)-)))).)).))). ( -32.90) >consensus GACAGGCCUCGUCUCACCUCAAGACAUUCCCAGCCCGGAUAAAUCACACAGCGAUGAAUUUGCAUAACUCCCGGUGCAGAUCGGUCGAGUAAUUUGUUCUGCUUAAGAGGAAACUAGUUU (((((.((((((((.......))))......(((..((((((((.((....((((..((((((((........))))))))..)))).)).)))))))).)))...))))...)).))). (-28.86 = -29.30 + 0.44)

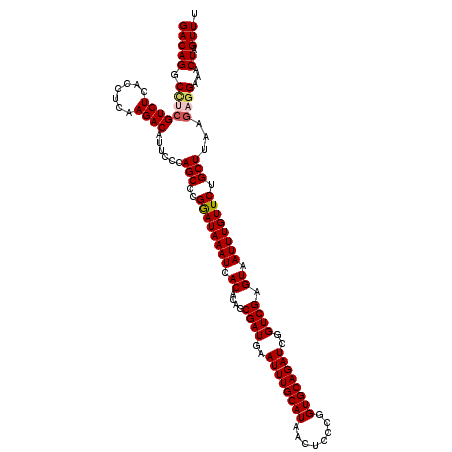
| Location | 21,738,918 – 21,739,038 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -36.06 |
| Energy contribution | -36.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21738918 120 - 23771897 CUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAUCCGUCGGCGUCCUGGCAAGGACAAUAGACGGGCCGACCGUUUCAGUUGCCUCACUUUAUGCGAG ((((....(((((((((.(.(((((((((.....))))))))).)))))))..(..(((((...(((((....)))))....)))))..)..)))..))))....(((.(.....).))) ( -44.30) >DroSec_CAF1 20814 119 - 1 CUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCU-GCAAGGACAAUAACCGGGCUGUCCGUUUCAGUUGCCUCACUUUAUGCGAG ..(((((....((((((.(.(((((((((.....))))))))).)))))))..)))))...((((..((-(...(((((..........)))))....))).)))).............. ( -44.20) >DroSim_CAF1 25706 119 - 1 CUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCU-GCAAGGACAAUAACCGGGCUGCCCGUUUCAGUUGCCUCACUUUAUGCGAG ..(((((....((((((.(.(((((((((.....))))))))).)))))))..)))))....((((...-(..(((.((((((.((((...)))).))..)))))))..)...))))... ( -43.40) >DroEre_CAF1 6889 120 - 1 CUCGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCAGUCGGCAUCCUGGCAAGGACAAUGAACGGGCUGCUCGUUUCAUUUGCCUCACUUUAUGCGGG ......((((.((((((.(.(((((((((.....))))))))).)))))))..((((((((.(((((((....))))...))..).))))))))......................)))) ( -44.20) >DroYak_CAF1 78463 111 - 1 CUCGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCAGUUGGCAUCCUGACAAGGACAACAGCCGGGCU---------AUUUGCCUCACUUUAUGCGAG (((((...(((((((((.(.(((((((((.....))))))))).))))))........((((...((((....)))))))).))))(((.---------....))).........))))) ( -42.40) >consensus CUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCUGGCAAGGACAAUAACCGGGCUGCCCGUUUCAGUUGCCUCACUUUAUGCGAG .((((((....((((((.(.(((((((((.....))))))))).)))))))..))))))...(((((((....))))........((((..............)))).......)))... (-36.06 = -36.50 + 0.44)

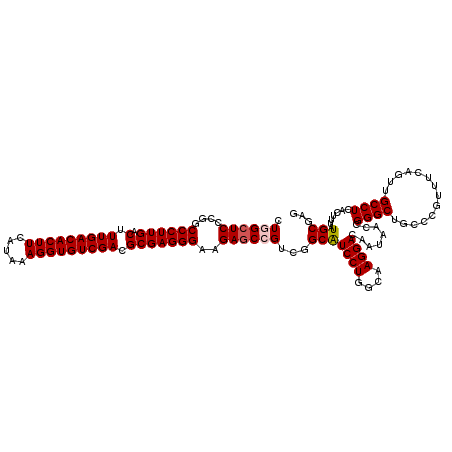
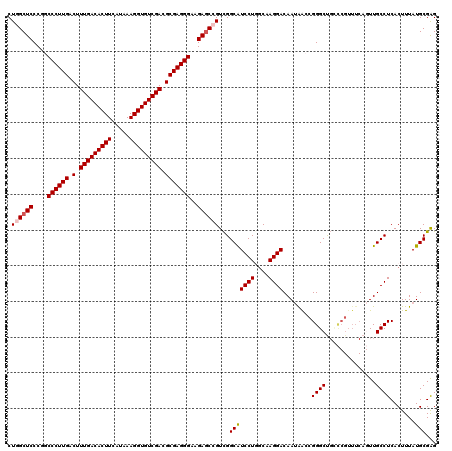
| Location | 21,738,958 – 21,739,078 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -34.24 |
| Energy contribution | -35.20 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
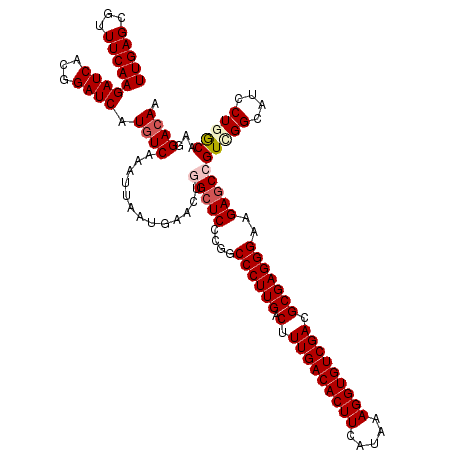
>3L_DroMel_CAF1 21738958 120 - 23771897 UUGAGCGUUUCAAGAUCACGGAUCAUGUCAAAUUAAUGAACUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAUCCGUCGGCGUCCUGGCAAGGACAA (((((...)))))(.(((((((((................((((...))))((((((.(.(((((((((.....))))))))).)))))))..))))))).)))(((((....))))).. ( -41.50) >DroSec_CAF1 20854 119 - 1 UUGAGCGUUUCAAGAUCACGGAUCAUGUCAAAUUAAUGAACUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCU-GCAAGGACAA (((((...)))))((((...)))).((((.......(((...(((((....((((((.(.(((((((((.....))))))))).)))))))..))))).)))((.....-))...)))). ( -38.10) >DroSim_CAF1 25746 119 - 1 UUGAGCGUUUCAAGAUCACGGAUCAUGUCAAAUUAAUGAACUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCU-GCAAGGACAA (((((...)))))((((...)))).((((.......(((...(((((....((((((.(.(((((((((.....))))))))).)))))))..))))).)))((.....-))...)))). ( -38.10) >DroEre_CAF1 6929 120 - 1 UUGAGCGUUUCAAGAUCACGGAUCAUGUCACAUUAAUGAACUCGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCAGUCGGCAUCCUGGCAAGGACAA ....((((..((.((((...)))).))..)).....(((.((.((((....((((((.(.(((((((((.....))))))))).)))))))..))))))))))).((((....))))... ( -38.60) >DroYak_CAF1 78494 120 - 1 UUGAGCCUUUCAAGAUCACGGAUCAUAUCAAAUUAAUGAACUCGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCAGUUGGCAUCCUGACAAGGACAA ....(((......((((...))))..............((((.((((....((((((.(.(((((((((.....))))))))).)))))))..))))))))))).((((....))))... ( -40.10) >consensus UUGAGCGUUUCAAGAUCACGGAUCAUGUCAAAUUAAUGAACUGGCUCCCGGCCCUUGACUUUGACACUUCAUAAAGGUGUCGACGCGAGGGAAGAGCCGUCGGCAUCCUGGCAAGGACAA (((((...)))))((((...)))).((((.............(((((....((((((.(.(((((((((.....))))))))).)))))))..)))))(((((....)))))...)))). (-34.24 = -35.20 + 0.96)

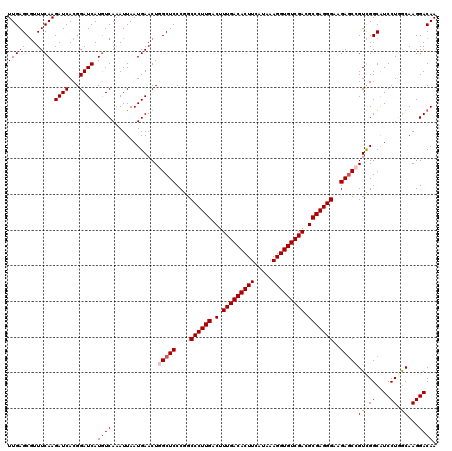
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:30 2006