
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,732,123 – 21,732,237 |
| Length | 114 |
| Max. P | 0.682974 |

| Location | 21,732,123 – 21,732,237 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -28.99 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
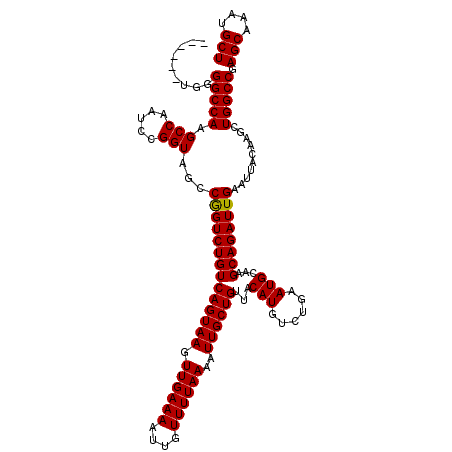
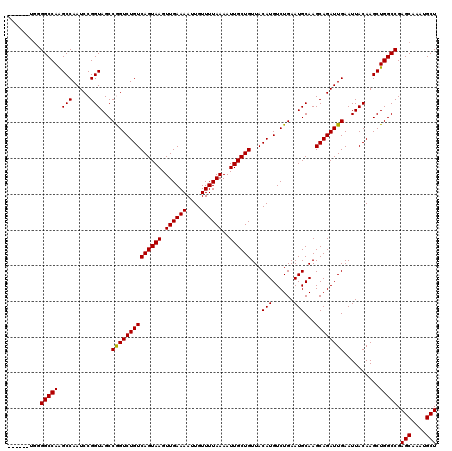
>3L_DroMel_CAF1 21732123 114 + 23771897 ------UGGGGCCAAGCCAAUCCGGUAGCCGGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ------...(((((.((((..((((...))))..)).((((((.((((((....))))))..))))))...((.(((((........)))))))........))))))).(((....))) ( -28.40) >DroPse_CAF1 42047 114 + 1 ------UGAGGCCAGGCCAAUCCGGUAGCCAGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ------...(((((((((.....)))...((((((((((((((.((((((....))))))..))))))...(((......)))...)))))))).........)))))).(((....))) ( -32.70) >DroSim_CAF1 18889 114 + 1 ------UGGGGCCAAGCCAAUCCGGUAGCCGGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ------...(((((.((((..((((...))))..)).((((((.((((((....))))))..))))))...((.(((((........)))))))........))))))).(((....))) ( -28.40) >DroPer_CAF1 40735 114 + 1 ------UGAGGCCAGGCCAAUCCGGUAGCCAGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ------...(((((((((.....)))...((((((((((((((.((((((....))))))..))))))...(((......)))...)))))))).........)))))).(((....))) ( -32.70) >DroEre_CAF1 119 111 + 1 ---------GGCCAAGCCAAUCCGGUAGCCGGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ---------(((((.((((..((((...))))..)).((((((.((((((....))))))..))))))...((.(((((........)))))))........))))))).(((....))) ( -28.40) >DroYak_CAF1 64980 120 + 1 GGGGCUUGGGGCCAAGCCAAUCCGGUAGCCGGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU ((((((((....))))))...))....(((((..(((((((((.((((((....))))))..))))))...((.(((((........)))))))....)))..)))))..(((....))) ( -34.40) >consensus ______UGGGGCCAAGCCAAUCCGGUAGCCGGUCUGUCAGUAAGUUGAAAAUUGUUUUAAAAUUGCUGUUACAUGUCUGAAUGCAAGCAGAUUGAAUUACAAGCUGGCCGAGCAAAUGCU .........(((((.(((.....)))...((((((((((((((.((((((....))))))..))))))...(((......)))...))))))))..........))))).(((....))) (-28.99 = -28.77 + -0.22)

| Location | 21,732,123 – 21,732,237 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
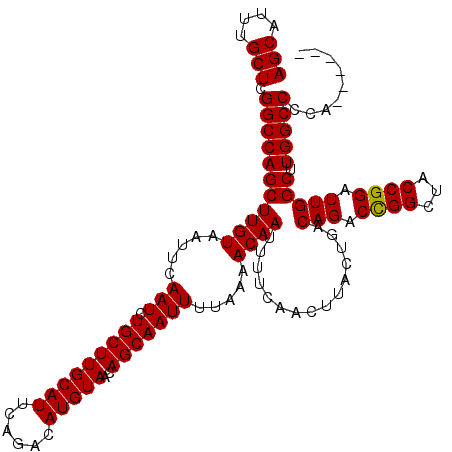
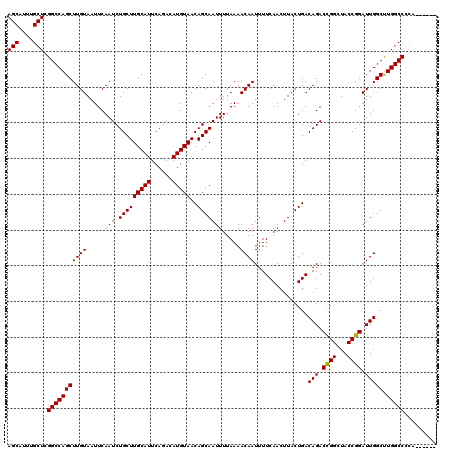
>3L_DroMel_CAF1 21732123 114 - 23771897 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACCGGCUACCGGAUUGGCUUGGCCCCA------ (((....))).(((((((((((.....(((.(((((((((......)))))..))))))).....))))...............(((.((((...)))).))))).)))))...------ ( -25.30) >DroPse_CAF1 42047 114 - 1 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACUGGCUACCGGAUUGGCCUGGCCUCA------ .......(((.((((((((.(((..(((.((((.((((((......))))))....................(((......))))))).))).))).)).)))))).)))....------ ( -24.40) >DroSim_CAF1 18889 114 - 1 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACCGGCUACCGGAUUGGCUUGGCCCCA------ (((....))).(((((((((((.....(((.(((((((((......)))))..))))))).....))))...............(((.((((...)))).))))).)))))...------ ( -25.30) >DroPer_CAF1 40735 114 - 1 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACUGGCUACCGGAUUGGCCUGGCCUCA------ .......(((.((((((((.(((..(((.((((.((((((......))))))....................(((......))))))).))).))).)).)))))).)))....------ ( -24.40) >DroEre_CAF1 119 111 - 1 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACCGGCUACCGGAUUGGCUUGGCC--------- (((....))).(((((((((((.....(((.(((((((((......)))))..))))))).....))))...............(((.((((...)))).))))).)))))--------- ( -24.50) >DroYak_CAF1 64980 120 - 1 AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACCGGCUACCGGAUUGGCUUGGCCCCAAGCCCC .((....))((((..((((.((.........(((((((((......)))))..))))...............(((......)))...)).)))).))))...((((((....)))))).. ( -26.10) >consensus AGCAUUUGCUCGGCCAGCUUGUAAUUCAAUCUGCUUGCAUUCAGACAUGUAACAGCAAUUUUAAAACAAUUUUCAACUUACUGACAGACCGGCUACCGGAUUGGCUUGGCCCCA______ (((....))).(((((((((((.....(((.(((((((((......)))))..))))))).....))))...............(((.((((...)))).))))).)))))......... (-24.79 = -24.57 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:19 2006