
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,731,584 – 21,731,717 |
| Length | 133 |
| Max. P | 0.835754 |

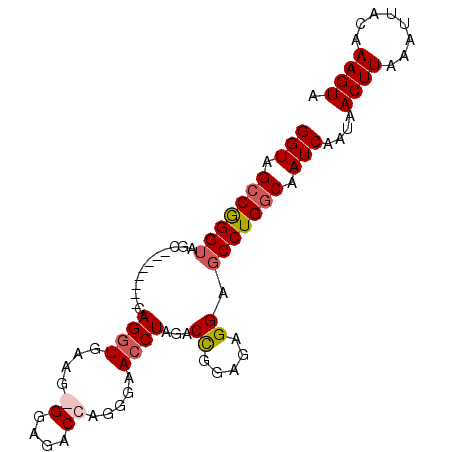
| Location | 21,731,584 – 21,731,677 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.33 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21731584 93 + 23771897 GGUAGCCGGGUAGCCAGGUAGCCAGGUGAAGGGGAGACCAGGAAACCUAGACCGGAGAGGAGCCUGGCAAUCAAUAACUUAAAUUACAAAGUA (((.(((((((..((.(((....((((....((....)).....))))..))).....)).))))))).)))....((((........)))). ( -29.60) >DroSec_CAF1 13292 84 + 1 GGUAGCCGGGUAGC--------CAGGUGAAG-GGAGACCAGGGAACCUAGACCGGAGAGGAGCCUGGCAAUCAAUAACUUAAAUUACAAAGUA (((.(((((((...--------.((((...(-(....)).....))))...((.....)).))))))).)))....((((........)))). ( -25.80) >DroSim_CAF1 18359 84 + 1 GGUAGCCGGGUAGC--------CAGGUGAAG-GGAGACCAGGGAACCUAGACCGGAGAGGAGCCUGGCAAUCAAUAACUUAAAUUACAAAGUA (((.(((((((...--------.((((...(-(....)).....))))...((.....)).))))))).)))....((((........)))). ( -25.80) >DroYak_CAF1 64465 85 + 1 GGUUGGCAGGGAGC--------CAGGUGAAGGGGAAUCCAGGGAAGCUAGACUGGCGAGGAGCCUGGCAAUCAAUAACUUAAAUUACAAAGUA (((((.((((..((--------(((.......((..((....))..))...)))))......)))).)))))....((((........)))). ( -23.60) >consensus GGUAGCCGGGUAGC________CAGGUGAAG_GGAGACCAGGGAACCUAGACCGGAGAGGAGCCUGGCAAUCAAUAACUUAAAUUACAAAGUA (((.(((((((............((((....((....)).....))))...((.....)).))))))).)))....((((........)))). (-18.33 = -19.20 + 0.87)


| Location | 21,731,618 – 21,731,717 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -22.45 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21731618 99 - 23771897 CAGCGAAGCUGCCUCAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAGGCUCCUCUCCGGUCUAGGUUU--------------CCUG------GUCU- (((((..((((...)))).))))).((..(((((.(((((((....(((((....)))))))))))))))))...))...((.(((((...--------------))))------).))- ( -31.30) >DroPse_CAF1 41534 120 - 1 AAGCGAAGCUGCUACAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAAGCUCCUCUCUAGUCUAGGCGCGGGCUCAUCUCUCUCCUAACUCCCAUCUC .((((..((((...)))).))))((((..((((((.((((.((((........)))).))))...(((.((((.......)).)).)))))))))))))..................... ( -27.00) >DroSec_CAF1 13317 99 - 1 CAGCGAAGCUGCCUCAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAGGCUCCUCUCCGGUCUAGGUUC--------------CCUG------GUCU- (((((..((((...)))).))))).((..(((((.(((((((....(((((....)))))))))))))))))...))...((.(((((...--------------))))------).))- ( -31.30) >DroSim_CAF1 18384 99 - 1 CAGCGAAGCUGCCUCAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAGGCUCCUCUCCGGUCUAGGUUC--------------CCUG------GUCU- (((((..((((...)))).))))).((..(((((.(((((((....(((((....)))))))))))))))))...))...((.(((((...--------------))))------).))- ( -31.30) >DroPer_CAF1 40223 119 - 1 AAGCGAAGCUGCUACAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAAGCUCCUCUCUAGUCUAGGCGCGGGCUCAUCUCUCUCCUAACUCCCAUCU- .((((..((((...)))).))))((((..((((((.((((.((((........)))).))))...(((.((((.......)).)).)))))))))))))....................- ( -27.00) >DroYak_CAF1 64491 99 - 1 CAGCGAAGCUGCUUCAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAGGCUCCUCGCCAGUCUAGCUUC--------------CCUG------GAUU- (((.((((((((....))..(((((((..(((((.(((((((....(((((....)))))))))))))))))...))).))))..))))))--------------.)))------....- ( -35.20) >consensus CAGCGAAGCUGCCUCAGCGUGCUGUGAAUGCCUGUGCAAUUACUUUGUAAUUUAAGUUAUUGAUUGCCAGGCUCCUCUCCAGUCUAGGUUC______________CCUG______GUCU_ (((((..((((...)))).))))).((..(((((.(((((((....(((((....)))))))))))))))))...))........................................... (-22.45 = -23.12 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:17 2006