
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,727,092 – 21,727,271 |
| Length | 179 |
| Max. P | 0.999694 |

| Location | 21,727,092 – 21,727,191 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21727092 99 + 23771897 UUUGGUUUUGCGUUUUUUCCUACUCCUCGC---------------UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC ..(((((((((........(((((((....---------------.)))))))...------...((((....)))).((((...(((......)))...)))).)))..)))))).... ( -24.70) >DroPse_CAF1 35899 116 + 1 GCCAGCU----CUCUUCAUCUAUUUCUCAACUUUUUUUAACCUUUUCCAGUUGCCACUGCAGCUGAUCACUUGUCAUAGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC (((....----....................................(((((((....))))))).............((((...(((......)))...)))))))............. ( -22.60) >DroSec_CAF1 8708 99 + 1 UUUGGUUUUGCGUUUUUUCCAACUUCUCAC---------------UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC ..((((((((((((......))).......---------------((((((((..(------((((.(((((......))))).)))))))))))))........)))..)))))).... ( -22.70) >DroSim_CAF1 13748 99 + 1 UUUGGUUUUGCGUUUUUUCCAACUUCUCAC---------------UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC ..((((((((((((......))).......---------------((((((((..(------((((.(((((......))))).)))))))))))))........)))..)))))).... ( -22.70) >DroPer_CAF1 34471 116 + 1 GCCAGCU----CUCUUCAUCUAUUUCUCAACUUUUUUUAACCUUUUCCAGUUGCCACUGCAGCUGAUCACUUGUCAUAGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC (((....----....................................(((((((....))))))).............((((...(((......)))...)))))))............. ( -22.60) >DroYak_CAF1 59476 94 + 1 UUUGGCU----GUCUUUUGCUACUACUCGG---------------UGGAGUGGCUA------CUGU-GAGUUGUCAAGGAGUUUAUGGUUCAUUCCACCUACUCGGCAAUAAAUCAUGCC ...((((----(....(((((.......((---------------((((((((..(------((((-((..(.......)..))))))))))))))))).....))))).....)).))) ( -26.70) >consensus UUUGGCU____GUCUUUUCCUACUUCUCAC_______________UGGAGUGGCUA______CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCC .............................................(((((((((((......((.((((....))))))......))).)))))))).......((((........)))) (-12.93 = -13.40 + 0.47)



| Location | 21,727,092 – 21,727,191 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21727092 99 - 23771897 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCCUUGACAAGUCAACAG------UAGCCACUCCA---------------GCGAGGAGUAGGAAAAAACGCAAAACCAAA ((((........))))((((..((((......))))..)))).((((....))))...------...(((((((.---------------....))))).)).................. ( -24.80) >DroPse_CAF1 35899 116 - 1 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCUAUGACAAGUGAUCAGCUGCAGUGGCAACUGGAAAAGGUUAAAAAAAGUUGAGAAAUAGAUGAAGAG----AGCUGGC ((((........))))((((..((((......))))..))))............((((((.((((....))))................(((.(....).)))......----)))))). ( -25.20) >DroSec_CAF1 8708 99 - 1 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCCUUGACAAGUCAACAG------UAGCCACUCCA---------------GUGAGAAGUUGGAAAAAACGCAAAACCAAA ((((........))))((((..((((......))))..)))).((((....))))...------..(((((....---------------)))....(((......)))))......... ( -20.60) >DroSim_CAF1 13748 99 - 1 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCCUUGACAAGUCAACAG------UAGCCACUCCA---------------GUGAGAAGUUGGAAAAAACGCAAAACCAAA ((((........))))((((..((((......))))..)))).((((....))))...------..(((((....---------------)))....(((......)))))......... ( -20.60) >DroPer_CAF1 34471 116 - 1 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCUAUGACAAGUGAUCAGCUGCAGUGGCAACUGGAAAAGGUUAAAAAAAGUUGAGAAAUAGAUGAAGAG----AGCUGGC ((((........))))((((..((((......))))..))))............((((((.((((....))))................(((.(....).)))......----)))))). ( -25.20) >DroYak_CAF1 59476 94 - 1 GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGAACCAUAAACUCCUUGACAACUC-ACAG------UAGCCACUCCA---------------CCGAGUAGUAGCAAAAGAC----AGCCAAA (((.((.(((.((((.((((..((((......))))..))))...........-....------..((.((((..---------------..)))).)).)))).))))----))))... ( -22.70) >consensus GGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAACUCCUUGACAAGUCAACAG______UAGCCACUCCA_______________GUGAGAAGUAGGAAAAAAC____AACCAAA ((((........))))((((..((((......))))..)))).............................................................................. (-14.73 = -14.73 + -0.00)

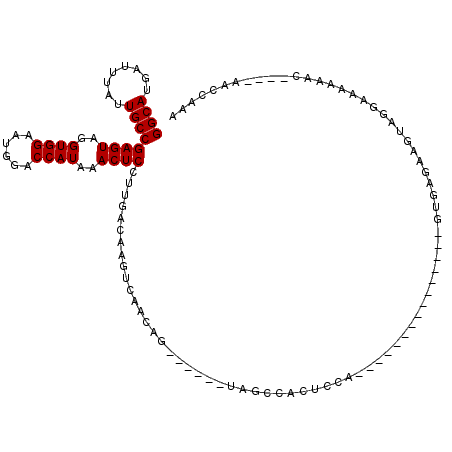
| Location | 21,727,122 – 21,727,231 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21727122 109 + 23771897 -----UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACC -----..(((((((.(------(..((((....)))).((((...(((......)))...))))((((........)))).........)).)))))))..................... ( -28.50) >DroPse_CAF1 35935 120 + 1 CCUUUUCCAGUUGCCACUGCAGCUGAUCACUUGUCAUAGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCCAACAACC ...(((((((((((....))))))).......(.((((((((...(((......)))...))))((((........))))......)))).).............))))........... ( -25.50) >DroSec_CAF1 8738 109 + 1 -----UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACC -----..(((((((.(------(..((((....)))).((((...(((......)))...))))((((........)))).........)).)))))))..................... ( -28.50) >DroSim_CAF1 13778 109 + 1 -----UGGAGUGGCUA------CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACC -----..(((((((.(------(..((((....)))).((((...(((......)))...))))((((........)))).........)).)))))))..................... ( -28.50) >DroPer_CAF1 34507 120 + 1 CCUUUUCCAGUUGCCACUGCAGCUGAUCACUUGUCAUAGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCCAACAACC ...(((((((((((....))))))).......(.((((((((...(((......)))...))))((((........))))......)))).).............))))........... ( -25.50) >DroYak_CAF1 59502 108 + 1 -----UGGAGUGGCUA------CUGU-GAGUUGUCAAGGAGUUUAUGGUUCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACC -----..(((((((.(------(.((-((((((.....((((...(((......)))...))))((((........)))).)))))))))).)))))))..................... ( -31.40) >consensus _____UGGAGUGGCUA______CUGUUGACUUGUCAAGGAGUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACC .......(((((((........................((((...(((......)))...))))((((........))))............)))))))..................... (-22.23 = -22.90 + 0.67)



| Location | 21,727,151 – 21,727,271 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.45 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.18 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21727151 120 + 23771897 GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACCGAAUAUGGCGGCACGGUGCCAUAAUUUCAUAGCCAAGUUA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))................ ( -25.00) >DroPse_CAF1 35975 119 + 1 GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCCAACAACCGAAUAUGGCGAUACGGUGCCAUAAUUUCAUAGCCA-GUAA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))...........-.... ( -23.70) >DroSec_CAF1 8767 120 + 1 GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACCGAAUAUGGCGGCACGGUGCCAUAAUUUCAUAGCCAAGUUA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))................ ( -25.00) >DroSim_CAF1 13807 120 + 1 GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACCGAAUAUGGCGGCACGGUGCCAUAAUUUCAUAGCCAAGUUA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))................ ( -25.00) >DroPer_CAF1 34547 119 + 1 GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCCAACAACCGAAUAUGGCGAUACGGUGCCAUAAUUUCAUAGCCA-GUAA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))...........-.... ( -23.70) >DroYak_CAF1 59530 120 + 1 GUUUAUGGUUCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACCGAAUAUGGCAGCACGGUGCCAUAAUUUCAUAGCCAAGUUA .....(((((..............((((........))))........(((((....((......)).....)))))...((((((((((......)))))))..)))..)))))..... ( -21.40) >consensus GUUUAUGGUCCAUUCCACCUACUCGGCAAUAAAUCAUGCCUUAAUUUAUGUCGCCACUCAAUAACGAAACCCUGACAACCGAAUAUGGCGGCACGGUGCCAUAAUUUCAUAGCCAAGUUA ..(((((((((.............((((........))))........((((((((.((.....................))...)))))))).)).)))))))................ (-23.86 = -23.45 + -0.41)



| Location | 21,727,151 – 21,727,271 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -35.40 |
| Energy contribution | -34.77 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21727151 120 - 23771897 UAACUUGGCUAUGAAAUUAUGGCACCGUGCCGCCAUAUUCGGUUGUCAGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ................((((((..((((.(((((.(((((.(((((((....((((....))))))))))).........((((........)))))))))))))).)))).)))))).. ( -38.30) >DroPse_CAF1 35975 119 - 1 UUAC-UGGCUAUGAAAUUAUGGCACCGUAUCGCCAUAUUCGGUUGUUGGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ....-...........((((((..((((.(((((.((((((((..........(((((((...))))))).((((((((.....)))))))).))))))))))))).)))).)))))).. ( -34.10) >DroSec_CAF1 8767 120 - 1 UAACUUGGCUAUGAAAUUAUGGCACCGUGCCGCCAUAUUCGGUUGUCAGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ................((((((..((((.(((((.(((((.(((((((....((((....))))))))))).........((((........)))))))))))))).)))).)))))).. ( -38.30) >DroSim_CAF1 13807 120 - 1 UAACUUGGCUAUGAAAUUAUGGCACCGUGCCGCCAUAUUCGGUUGUCAGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ................((((((..((((.(((((.(((((.(((((((....((((....))))))))))).........((((........)))))))))))))).)))).)))))).. ( -38.30) >DroPer_CAF1 34547 119 - 1 UUAC-UGGCUAUGAAAUUAUGGCACCGUAUCGCCAUAUUCGGUUGUUGGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ....-...........((((((..((((.(((((.((((((((..........(((((((...))))))).((((((((.....)))))))).))))))))))))).)))).)))))).. ( -34.10) >DroYak_CAF1 59530 120 - 1 UAACUUGGCUAUGAAAUUAUGGCACCGUGCUGCCAUAUUCGGUUGUCAGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGAACCAUAAAC ..(((((((....((((((((.((((.(((.(((......))).).)).))).(((((((...)))))))..........).))))))))...)))))))..((((......)))).... ( -36.60) >consensus UAACUUGGCUAUGAAAUUAUGGCACCGUGCCGCCAUAUUCGGUUGUCAGGGUUUCGUUAUUGAGUGGCGACAUAAAUUAAGGCAUGAUUUAUUGCCGAGUAGGUGGAAUGGACCAUAAAC ................((((((..((((.(((((.(((((.(((((((....((((....))))))))))).........((((........)))))))))))))).)))).)))))).. (-35.40 = -34.77 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:11 2006