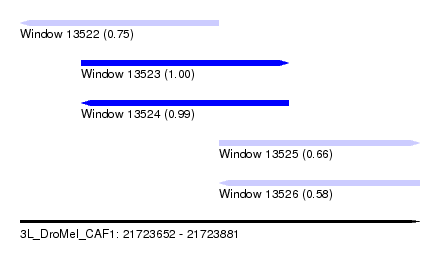
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,723,652 – 21,723,881 |
| Length | 229 |
| Max. P | 0.999859 |

| Location | 21,723,652 – 21,723,766 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -42.49 |
| Energy contribution | -42.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21723652 114 - 23771897 AAUCGAUGGCAGUGCCGCCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUCAGCUUCAGCUGCAGCCUUAGCUUGCGAUUGCGAUU----- (((((..(((.(((((..(((......))).))))).........(((..((((...))))((((((((((((....)))))..))))))).)))...)))..)))))......----- ( -41.80) >DroSec_CAF1 5564 119 - 1 AAUCGAUGGCAGUGCCGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUCAGCUUCAGCUGCAGCCUUAGCCUGCGAUUGCGAUUGCCGC ((((((((.(.(((((.((((......)))))))))..(.....)).))))))))....((((((((((((((..((.((((....)))).))..)))))).((....)))))))))). ( -47.20) >DroSim_CAF1 10583 119 - 1 AAUCGAUGGCAGUGCCGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUCAGCUUCAGCUGCAGCCUUAGCCUGCGAUUGCGAUUGCCGC ((((((((.(.(((((.((((......)))))))))..(.....)).))))))))....((((((((((((((..((.((((....)))).))..)))))).((....)))))))))). ( -47.20) >consensus AAUCGAUGGCAGUGCCGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUCAGCUUCAGCUGCAGCCUUAGCCUGCGAUUGCGAUUGCCGC (((((..(((.(((((.((((......))))))))).........(((..((((...))))((((((((((((....)))))..))))))).)))...)))..)))))........... (-42.49 = -42.60 + 0.11)



| Location | 21,723,687 – 21,723,806 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.14 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21723687 119 + 23771897 GAAGUACCAGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGGCG-GCACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC ...(((.(((....)))......((((((((((.((((..((....((((........))))))-)))).).)))))))))...)))......(((((((((.....).))))))))... ( -35.30) >DroPse_CAF1 32378 112 + 1 --------AGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGACGGGCAUCGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC --------...........(((.(((((((((.((......((((.((((((......)))).)))))))).))))))))).)))........(((((((((.....).))))))))... ( -33.60) >DroSec_CAF1 5604 119 + 1 GAAGUACCAGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGACG-GCACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC ...(((.(((....)))......((((((((((.((((..(((....(((........))))))-)))).).)))))))))...)))......(((((((((.....).))))))))... ( -33.70) >DroSim_CAF1 10623 119 + 1 GAAGUACCAGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGACG-GCACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC ...(((.(((....)))......((((((((((.((((..(((....(((........))))))-)))).).)))))))))...)))......(((((((((.....).))))))))... ( -33.70) >DroPer_CAF1 31092 112 + 1 --------AGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGACGGGCAUCGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC --------...........(((.(((((((((.((......((((.((((((......)))).)))))))).))))))))).)))........(((((((((.....).))))))))... ( -33.60) >consensus GAAGUACCAGCCAACUGCCGAUCAAAUCGAUGCCGUGUAUCGUGUACCUCUCCAUCCUGAGACG_GCACUGCCAUCGAUUUAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAAC ...................(((.((((((((((.((((....(((..(((........)))))).)))).).))))))))).)))........(((((((((.....).))))))))... (-32.54 = -32.14 + -0.40)



| Location | 21,723,687 – 21,723,806 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -36.36 |
| Energy contribution | -36.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21723687 119 - 23771897 GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCAGUGC-CGCCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUC ...((((((((........))))))))......(((((((((((((((.(.((((-(..(((......))).)))))..(.....)).))))))))))))((((.....)))).)))... ( -42.60) >DroPse_CAF1 32378 112 - 1 GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCGAUGCCCGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCU-------- ....(((((((........)))))))(((..((((.((((((((((((.((....(((((....)))))....(((.....))).)).))))))))))))..))))..))).-------- ( -41.70) >DroSec_CAF1 5604 119 - 1 GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCAGUGC-CGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUC ...((((((((........))))))))......(((((((((((((((.(.((((-(.((((......)))))))))..(.....)).))))))))))))((((.....)))).)))... ( -43.90) >DroSim_CAF1 10623 119 - 1 GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCAGUGC-CGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUC ...((((((((........))))))))......(((((((((((((((.(.((((-(.((((......)))))))))..(.....)).))))))))))))((((.....)))).)))... ( -43.90) >DroPer_CAF1 31092 112 - 1 GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCGAUGCCCGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCU-------- ....(((((((........)))))))(((..((((.((((((((((((.((....(((((....)))))....(((.....))).)).))))))))))))..))))..))).-------- ( -41.70) >consensus GUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUAAAUCGAUGGCAGUGC_CGUCUCAGGAUGGAGAGGUACACGAUACACGGCAUCGAUUUGAUCGGCAGUUGGCUGGUACUUC ....(((((((........)))))))(((..((((.((((((((((((.(.((((.(.((((......)))))))))..(.....)).))))))))))))..))))..)))......... (-36.36 = -36.96 + 0.60)



| Location | 21,723,766 – 21,723,881 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 97.40 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -23.73 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21723766 115 + 23771897 UAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAACCAGCUGAA-CCGAUCGAAAAACAGGAAAGGGGCCUAAUGC .............(((((((((.....).))))))))..((((.((...(((((...((...))...(.(((((......))))-).)............)))))..))..)))). ( -27.50) >DroSec_CAF1 5683 115 + 1 UAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCGAACAGUUCAACCAGCUGAA-CCGAUCGAAAAACAGGAAAGGGGCCUAAUGC .............(((((((((.....).))))))))..((((.((...(((((.........(((.(.(((((......))))-).).)))........)))))..))..)))). ( -30.43) >DroSim_CAF1 10702 115 + 1 UAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAACCAGCUGAA-CCGAUCGAAAAACAGGAAAGGGGCCUAAUGC .............(((((((((.....).))))))))..((((.((...(((((...((...))...(.(((((......))))-).)............)))))..))..)))). ( -27.50) >DroYak_CAF1 56287 116 + 1 UAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCAAAGCAAAGCAAACAGUUCAACCAGCUGAAGCCGAUCGAAAAACAGGAAAGGAGGCUAAAGC ..............((((((((.....).)))))))((((.(((((................)))))..)))).....(((..((((..((.............)).))))..))) ( -26.71) >consensus UAAUUACAACAAACGCGUCAGCAAAUUGCCUGACGCGAACAUUUGCACACUUUCGAAGCAAAGCAAACAGUUCAACCAGCUGAA_CCGAUCGAAAAACAGGAAAGGGGCCUAAUGC .............(((((((((.....).))))))))..((((.((...(((((...((...))...(((((.....)))))..................)))))..))..)))). (-23.73 = -24.23 + 0.50)


| Location | 21,723,766 – 21,723,881 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.40 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -30.01 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21723766 115 - 23771897 GCAUUAGGCCCCUUUCCUGUUUUUCGAUCGG-UUCAGCUGGUUGAACUGUUUGCUUUGCUUCGAAAGUGUGCAAAUGUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUA (((((..(((.(((((..((....(((.(((-(((((....)))))))).)))....))...))))).).)).))))).((((((((........))))))))............. ( -33.90) >DroSec_CAF1 5683 115 - 1 GCAUUAGGCCCCUUUCCUGUUUUUCGAUCGG-UUCAGCUGGUUGAACUGUUCGCUUUGCUUCGAAAGUGUGCAAAUGUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUA (((((..(((.(((((..((....(((.(((-(((((....)))))))).)))....))...))))).).)).))))).((((((((........))))))))............. ( -36.00) >DroSim_CAF1 10702 115 - 1 GCAUUAGGCCCCUUUCCUGUUUUUCGAUCGG-UUCAGCUGGUUGAACUGUUUGCUUUGCUUCGAAAGUGUGCAAAUGUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUA (((((..(((.(((((..((....(((.(((-(((((....)))))))).)))....))...))))).).)).))))).((((((((........))))))))............. ( -33.90) >DroYak_CAF1 56287 116 - 1 GCUUUAGCCUCCUUUCCUGUUUUUCGAUCGGCUUCAGCUGGUUGAACUGUUUGCUUUGCUUUGAAAGUGUGCAAAUGUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUA ((....))...(((((......((((((((((....))))))))))......((...))...)))))..(((((.....((((((((........))))))))....))))).... ( -32.70) >consensus GCAUUAGGCCCCUUUCCUGUUUUUCGAUCGG_UUCAGCUGGUUGAACUGUUUGCUUUGCUUCGAAAGUGUGCAAAUGUUCGCGUCAGGCAAUUUGCUGACGCGUUUGUUGUAAUUA ...........(((((..((....(((.(((.(((((....)))))))).)))....))...)))))..(((((.....((((((((........))))))))....))))).... (-30.01 = -29.82 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:11:02 2006