
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,721,934 – 21,722,078 |
| Length | 144 |
| Max. P | 0.781318 |

| Location | 21,721,934 – 21,722,038 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -27.35 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
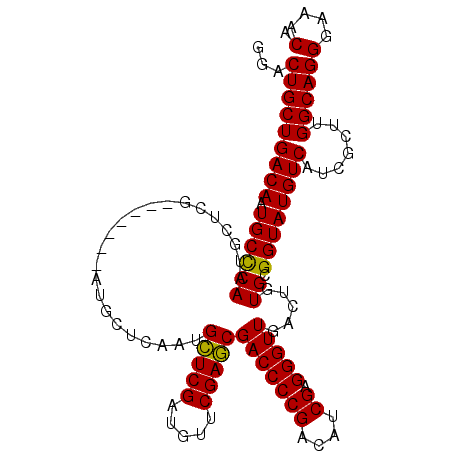
>3L_DroMel_CAF1 21721934 104 - 23771897 UCGAUGUCGGGGUCGCUCGAACAUCGAGCAUUGAGCAU-------CGAGCAUUGGGCAUUGUCAGCAGUCCCGUUCAACAAAGGGACAAUGUUUCUCGAUAAAUAUUGUUG (((((((((((....)))).)))))))..((((((...-------.((((((((((.((((....)))))))((((.......)))))))))))))))))........... ( -32.50) >DroSec_CAF1 3882 111 - 1 UCGAUGUCGGGGUCGCUCGAACAUCGAGCAUUGAGCAUCGAGCAUCGAGCAUUGGGCAUUGUCAGCAGUCCCGUUCAACAAAGGGACAAUGUUUCUCGAUAAAUAUUGUUG .(((((((.((...((((((...((((((.....)).))))...)))))).)).))))))).((((.(((((..........)))))(((((((......))))))))))) ( -37.50) >DroSim_CAF1 8571 111 - 1 UCGAUGUCGGGGUCGCUCGAACAUCGAGCAUUGAGCAUCGAGCAUCGAGCAUUGGGCAUUGUCAGCAGUCCCGUUCAACAAAGGGACAAUGUUUCUCGAUAAAUAUUGUUG .(((((((.((...((((((...((((((.....)).))))...)))))).)).))))))).((((.(((((..........)))))(((((((......))))))))))) ( -37.50) >DroYak_CAF1 54452 97 - 1 UCGAUGUCGGGGUCGUUCGAACAUCGAACAUUGGGCA--------------UUGAGCAUUGUCAGCAGUGCCGUGCAACAAAGGGACAAUGUUUCUCGAUAAAUAUUGUUG ((((((((.((...(((((.....))))).)).))))--------------))))((((((....)))))).............((((((((((......)))))))))). ( -30.40) >consensus UCGAUGUCGGGGUCGCUCGAACAUCGAGCAUUGAGCAU_______CGAGCAUUGGGCAUUGUCAGCAGUCCCGUUCAACAAAGGGACAAUGUUUCUCGAUAAAUAUUGUUG (((((((((((....)))).)))))))...((((((.(((.....))).....(((.((((....)))))))))))))......((((((((((......)))))))))). (-27.35 = -28.60 + 1.25)



| Location | 21,721,974 – 21,722,078 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.36 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21721974 104 + 23771897 GGACUGCUGACAAUGCCCAAUGCUCG-------AUGCUCAAUGCUCGAUGUUCGAGCGACCCCGACAUCGAGGGUUGACUGUGCGGUAUGUCAUCGCUUGGCAGGGAAACA ...((((..(.......((((.((((-------(((.((..((((((.....)))))).....))))))))).)))).....(((((.....))))))..))))(....). ( -38.40) >DroSec_CAF1 3922 111 + 1 GGACUGCUGACAAUGCCCAAUGCUCGAUGCUCGAUGCUCAAUGCUCGAUGUUCGAGCGACCCCGACAUCGAGGGUUGACUGUGCGGUAUGUCAUCGCUUGGCAGGGAAACA ...(((((((((.((((((..((((((.((((((.((.....)))))).))))))))(((((((....)).))))).....)).)))))))).......)))))(....). ( -42.51) >DroSim_CAF1 8611 111 + 1 GGACUGCUGACAAUGCCCAAUGCUCGAUGCUCGAUGCUCAAUGCUCGAUGUUCGAGCGACCCCGACAUCGAGGGUUGACUGUGCGGUAUGUCAUCGCUUGGCAGGGAAACA ...(((((((((.((((((..((((((.((((((.((.....)))))).))))))))(((((((....)).))))).....)).)))))))).......)))))(....). ( -42.51) >DroYak_CAF1 54492 97 + 1 GCACUGCUGACAAUGCUCAA--------------UGCCCAAUGUUCGAUGUUCGAACGACCCCGACAUCGAGGGUUGACUGUGCGGUAUGUCAUCGCUUGGCAGGGAAACA ...((((..((.(((....(--------------((((((..(((((.....)))))(((((((....)).))))).....)).)))))..))).).)..))))(....). ( -32.80) >consensus GGACUGCUGACAAUGCCCAAUGCUCG_______AUGCUCAAUGCUCGAUGUUCGAGCGACCCCGACAUCGAGGGUUGACUGUGCGGUAUGUCAUCGCUUGGCAGGGAAACA ...(((((((((.((((((.......................(((((.....)))))(((((((....)).))))).....)).)))))))).......)))))(....). (-32.92 = -32.36 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:57 2006