
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,718,761 – 21,718,881 |
| Length | 120 |
| Max. P | 0.633879 |

| Location | 21,718,761 – 21,718,881 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -25.08 |
| Energy contribution | -24.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
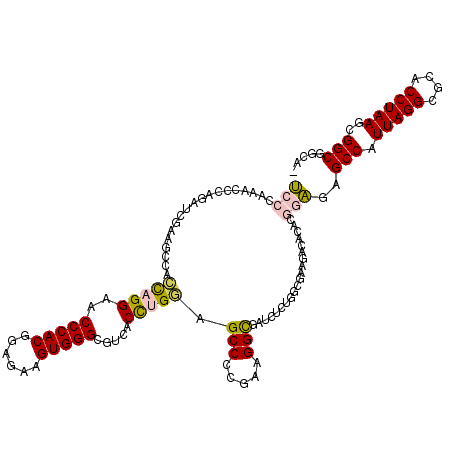
>3L_DroMel_CAF1 21718761 120 + 23771897 UCCCAAACCCAGAUCGAAGCCACCAGGAACCCACGGAGAAGUGGGCGUCACCUGGAGCCCCGAAGGUGAUCUCUGGCGAAGACACACGGAGAGCCAUUAGGCGCACCUAAGCGGCGGCCA ..................(((.(((((..(((((......))))).....))))).(((.(..(((((.((((((...........))))))(((....))).)))))..).)))))).. ( -46.30) >DroPse_CAF1 26821 93 + 1 C-------------CGCAGGCAAUCGGAACCCACCGAGAAGUGGGCGUCACCCGUUGCC-CAAAGGC------------AGACACACAUGGGGCCAUUAGGCGCACCUAAGCGGCAAGA- (-------------(((.(((....((..(((((......)))))..)).(((((((((-....)))------------).......)))))))).(((((....))))))))).....- ( -35.41) >DroSec_CAF1 708 119 + 1 UCCCAAACCCAGAUCGAAGCCACCAGGAACCCACGGAGAAGUGGGCGUCACUUGCAGCCCCGAAGGCGAUCUCUGGCGAGGACCCACGGAGAGCCAUUAGGCGCACCUAAGCGGCGGCA- ..................(((.((.((..(((((......))))).(((.(((((.(((.....)))(....)..))))))))))..))...(((.(((((....)))))..)))))).- ( -40.20) >DroSim_CAF1 4873 119 + 1 UCCCAAACCCAGAUCGAAGCCACCAGGAACCCACGGAGAAGUGGGCGUCACUUGGAGCCCCGAAGGCGAUCUCUGGCGAAGACCCACGGAGAGCCAUUAGGCGCACCUAAGCGGCGGCA- ..................(((.((.((..(((((......)))))..)).(((((.((((..(.(((..(((.(((.(....)))).)))..))).)..)).)).)).))).)).))).- ( -41.40) >DroPer_CAF1 25931 93 + 1 C-------------CGCAGGCAAUCGGAACCCACCGAGAAGUGGGCGUCACCCGUUGCC-CAAAGGC------------AGACACACAUGGGGCCAUUAGGCGCACCUAAGCGGCAAGA- (-------------(((.(((....((..(((((......)))))..)).(((((((((-....)))------------).......)))))))).(((((....))))))))).....- ( -35.41) >DroYak_CAF1 51319 119 + 1 UCCCAAACCCAUAUCGAAGCCACCAGGAACCCACGAAGAAGUGGGCGUCACCUGGAGCCCCGAAGGCGAACUCUAUCGAGGACCCACGGAGAGCCAUUAGGCGCACCUAAGCGGCGGGA- ((((....((...(((..((..(((((..(((((......))))).....))))).))..))).((.(..(((....)))..)))..))...(((.(((((....)))))..)))))))- ( -44.30) >consensus UCCCAAACCCAGAUCGAAGCCACCAGGAACCCACGGAGAAGUGGGCGUCACCUGGAGCCCCGAAGGCGAUCUCUGGCGAAGACACACGGAGAGCCAUUAGGCGCACCUAAGCGGCGGCA_ (((...................(((((..(((((......))))).....))))).(((.....)))....................)))..(((.(((((....)))))..)))..... (-25.08 = -24.88 + -0.19)

| Location | 21,718,761 – 21,718,881 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -29.16 |
| Energy contribution | -31.10 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21718761 120 - 23771897 UGGCCGCCGCUUAGGUGCGCCUAAUGGCUCUCCGUGUGUCUUCGCCAGAGAUCACCUUCGGGGCUCCAGGUGACGCCCACUUCUCCGUGGGUUCCUGGUGGCUUCGAUCUGGGUUUGGGA ...((((((.(((((....)))))))))((((.(.(((....)))).))))..(((((((((((((((((....((((((......)))))).))))).))))))))...))))..)).. ( -51.20) >DroPse_CAF1 26821 93 - 1 -UCUUGCCGCUUAGGUGCGCCUAAUGGCCCCAUGUGUGUCU------------GCCUUUG-GGCAACGGGUGACGCCCACUUCUCGGUGGGUUCCGAUUGCCUGCG-------------G -.....((((.((((..(((...(((....))))))..)))------------).....(-(((((((((....(((((((....))))))))))).)))))))))-------------) ( -38.10) >DroSec_CAF1 708 119 - 1 -UGCCGCCGCUUAGGUGCGCCUAAUGGCUCUCCGUGGGUCCUCGCCAGAGAUCGCCUUCGGGGCUGCAAGUGACGCCCACUUCUCCGUGGGUUCCUGGUGGCUUCGAUCUGGGUUUGGGA -.....((((...((.(.(((....))).).)))))).(((..(((..((((((((....))((..(.((.((.((((((......)))))))))).)..))..)))))).)))..))). ( -48.90) >DroSim_CAF1 4873 119 - 1 -UGCCGCCGCUUAGGUGCGCCUAAUGGCUCUCCGUGGGUCUUCGCCAGAGAUCGCCUUCGGGGCUCCAAGUGACGCCCACUUCUCCGUGGGUUCCUGGUGGCUUCGAUCUGGGUUUGGGA -.((((((((((.((.(((((....)))..((((.((((..((......))..)))).)))))).))))))((.((((((......))))))))..))))))(((((.......))))). ( -46.80) >DroPer_CAF1 25931 93 - 1 -UCUUGCCGCUUAGGUGCGCCUAAUGGCCCCAUGUGUGUCU------------GCCUUUG-GGCAACGGGUGACGCCCACUUCUCGGUGGGUUCCGAUUGCCUGCG-------------G -.....((((.((((..(((...(((....))))))..)))------------).....(-(((((((((....(((((((....))))))))))).)))))))))-------------) ( -38.10) >DroYak_CAF1 51319 119 - 1 -UCCCGCCGCUUAGGUGCGCCUAAUGGCUCUCCGUGGGUCCUCGAUAGAGUUCGCCUUCGGGGCUCCAGGUGACGCCCACUUCUUCGUGGGUUCCUGGUGGCUUCGAUAUGGGUUUGGGA -((((((((.(((((....)))))))))..(((((((((.(((....)))...))).(((((((((((((....((((((......)))))).))))).))))))))))))))...)))) ( -55.80) >consensus _UCCCGCCGCUUAGGUGCGCCUAAUGGCUCUCCGUGGGUCUUCGCCAGAGAUCGCCUUCGGGGCUCCAGGUGACGCCCACUUCUCCGUGGGUUCCUGGUGGCUUCGAUCUGGGUUUGGGA .....((((.(((((....))))))))).............................(((((((((((((....((((((......)))))).))))).))))))))............. (-29.16 = -31.10 + 1.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:55 2006