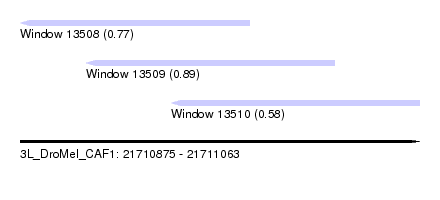
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,710,875 – 21,711,063 |
| Length | 188 |
| Max. P | 0.893235 |

| Location | 21,710,875 – 21,710,983 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21710875 108 - 23771897 CA--UCUCAUCUCGCC-UGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAA-ACCAACGCA-ACGGAGCG-------AGACAGCA ..--.....(((((((-((((((((........((((((..((........))...)))))).(((((....))))).........-..)))))).-.))).)))-------)))..... ( -28.80) >DroVir_CAF1 41237 103 - 1 AC--UCGCAGCUCUGCCUGCCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAG--------------UGUCCGGGCCAUGGGCAGC- ..--........((((((..(((((....)))))..........(((..(.(((((.......(((((....)))))........)--------------)))))..)))..)))))).- ( -29.76) >DroGri_CAF1 32786 118 - 1 UGCGUUGCAUCUCUGC-UGCCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAAAAGCAAAGAA-AUACAUCCAGCCCAUGGACAGCA .((.((((.....(((-((((((((....))))(........).))).))))...........(((((....)))))...........))))....-.....((((.....))))..)). ( -24.40) >DroSec_CAF1 50407 109 - 1 CA--UCUCAUCUCGCC-UGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAA-ACCAACACAGACGGAGCG-------AGACAGCA ..--.....(((((((-((((((((....)))))).......)))))..((............(((((....))))).........-.((.........)).)))-------)))..... ( -25.41) >DroSim_CAF1 45810 108 - 1 CA--UCUCAUCUCGCC-UGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAA-AA-ACCAACGCAGACGGAGCG-------AGACAGCA ..--.....(((((((-((((((((........((((((..((........))...)))))).(((((....)))))......-..-..))))))...))).)))-------)))..... ( -28.40) >DroMoj_CAF1 48905 101 - 1 UC--UGGCAACUGCGC-AGCCGUUGUCAACAAUGUAAUUUAUCAGGCAGGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAA--------------CGUCCAGCCCAUGG-CAGA- ..--(((((((.((..-.)).)))))))................((((((..(.....)..)))))).....((((..........--------------.).)))(((...))-)...- ( -23.90) >consensus CA__UCGCAUCUCGCC_UGCCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUAAAAAAA_ACCAACGCA_ACGGAGCC_______AGACAGCA .....................((((((......((((((..((........))...)))))).(((((....)))))....................................)))))). (-15.09 = -15.32 + 0.22)


| Location | 21,710,906 – 21,711,023 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 99.43 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21710906 117 - 23771897 GGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUA (((((((...(..................)...)))))))...........(((((((((((....)))))).......)))))................(((((....)))))... ( -29.38) >DroSec_CAF1 50439 117 - 1 GGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUA (((((((...(..................)...)))))))...........(((((((((((....)))))).......)))))................(((((....)))))... ( -29.38) >DroSim_CAF1 45841 117 - 1 GGCCGAGCUGCAAGCUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUA ((((((((.....))))............(.....)))))...........(((((((((((....)))))).......)))))................(((((....)))))... ( -30.41) >consensus GGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUUAUCAGGCAAGCGACAUAAUUACCUGCCACUUAUGGCAAUA (((((((...(..................)...)))))))...........(((((((((((....)))))).......)))))................(((((....)))))... (-29.38 = -29.38 + 0.00)

| Location | 21,710,946 – 21,711,063 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 98.86 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -25.93 |
| Energy contribution | -25.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21710946 117 - 23771897 CCGCCGCCACAUUACCGCUUUGCCAAAAACGAUUCCCCCGGGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUU ........(((((...((..(((((....(((.......((((((((...(..................)...))))))))........)))..)))))..))....)))))..... ( -25.93) >DroSec_CAF1 50479 117 - 1 CCGCCGCCACAUUACCGCUUUGCCAAAAUCGAUUCCCCCGGGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUU ........(((((...((..(((((....(((.......((((((((...(..................)...))))))))........)))..)))))..))....)))))..... ( -25.93) >DroSim_CAF1 45881 117 - 1 CCGCCGCCACAUUACCGCUUUGCCAAAAUCGAUUCCCCCGGGCCGAGCUGCAAGCUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUU ........(((((...((..(((((...(((((......(((..((((.....))))..)))....))))).....(((............))))))))..))....)))))..... ( -27.60) >consensus CCGCCGCCACAUUACCGCUUUGCCAAAAUCGAUUCCCCCGGGCCGAGCUGCAAACUCUCCCCACUCAUCGAAUCUCGGCCCAUCUCAUCUCGCCUGGCGUUGUCAACAAUGUAAUUU ........(((((...((..(((((....(((.......((((((((...(..................)...))))))))........)))..)))))..))....)))))..... (-25.93 = -25.93 + -0.00)

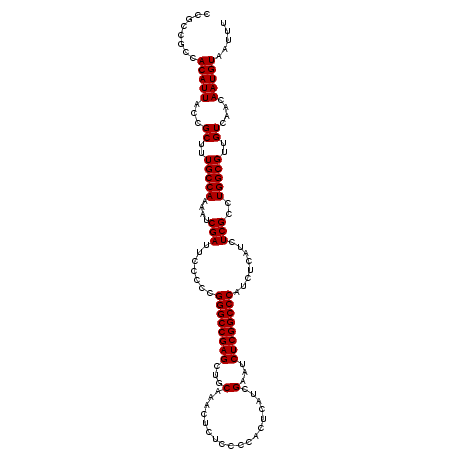
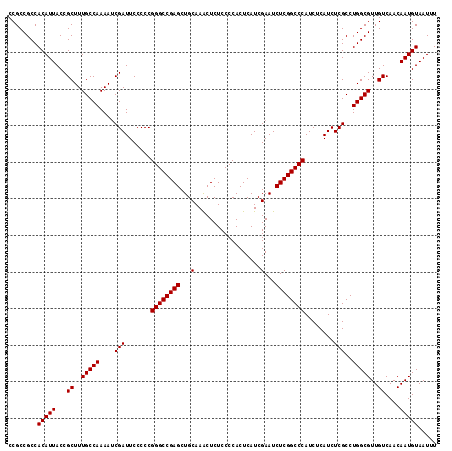
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:47 2006