
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,524,684 – 2,524,793 |
| Length | 109 |
| Max. P | 0.852792 |

| Location | 2,524,684 – 2,524,793 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -17.95 |
| Consensus MFE | -10.81 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.852792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
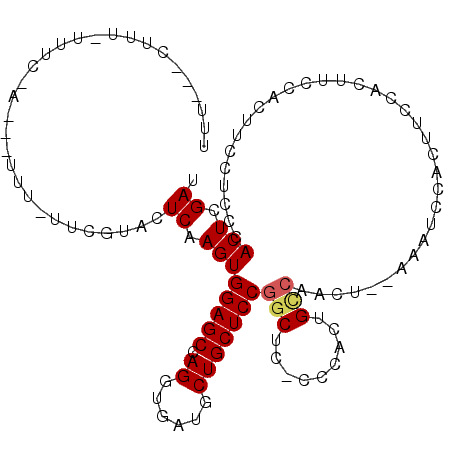
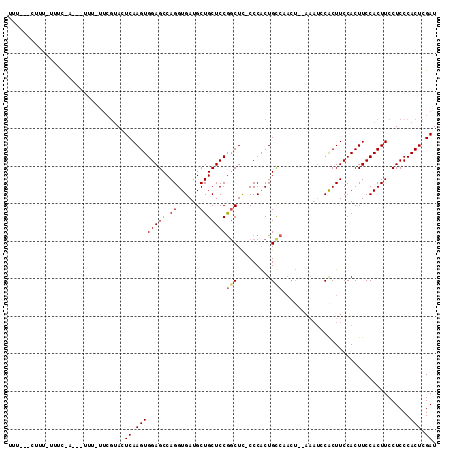
>3L_DroMel_CAF1 2524684 109 + 23771897 UUU---CUUU-UUUCAAACUUUUUUUCGUACUCAAGUGGAGCCAGGUGAUGCUGCUCCGGCUC-CCCACUGUCAACUGCAAAUCCGCUUCCACUUCCACUUCCUCCCACUCGAU ...---....-..............(((.....((((((((..(((((((..(((...(((..-......)))....))).)).)))))....)))))))).........))). ( -17.54) >DroSec_CAF1 147669 82 + 1 U------------UU-------------AACUCAAGUGGAGCCAGGUGAUGCUGCUCCGGCUC-CCCACUGCCAACUC------CACUUCCACUUCCACUUCCUCCCACUCGAU .------------..-------------.....((((((((...((.(......).))(((..-......)))..)))------)))))......................... ( -17.80) >DroYak_CAF1 147455 106 + 1 UUUCAGCUUUUUUUC-AG-CUUUGUUCGUAUUCAAGUGGAGCCAGGUGAUGCUGCUCCAACUUGCCCGCUGCA------AAAUCCACUUCCACUUCCACUUCCUCCCACUCGAU ..((.(((.......-))-)(((((.((....((((((((((.((......))))))).)))))..))..)))------))..............................)). ( -18.50) >consensus UUU___CUUU_UUUC_A___UUU_UUCGUACUCAAGUGGAGCCAGGUGAUGCUGCUCCGGCUC_CCCACUGCCAACU__AAAUCCACUUCCACUUCCACUUCCUCCCACUCGAU ...............................((.((((((((.((......)))))))(((.........)))..................................))).)). (-10.81 = -11.37 + 0.56)

| Location | 2,524,684 – 2,524,793 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -16.00 |
| Energy contribution | -15.34 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
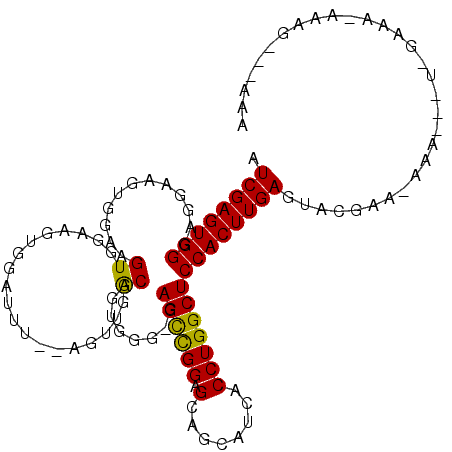
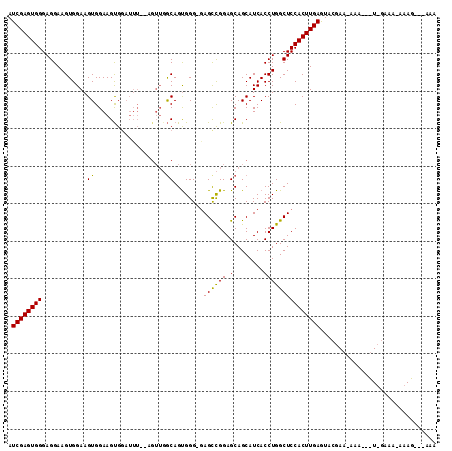
>3L_DroMel_CAF1 2524684 109 - 23771897 AUCGAGUGGGAGGAAGUGGAAGUGGAAGCGGAUUUGCAGUUGACAGUGGG-GAGCCGGAGCAGCAUCACCUGGCUCCACUUGAGUACGAAAAAAAGUUUGAAA-AAAG---AAA .((((((((((((..(..((..((....))..))..).((((.(..(((.-...)))..)))))....)))...)))))))))...((((......))))...-....---... ( -26.30) >DroSec_CAF1 147669 82 - 1 AUCGAGUGGGAGGAAGUGGAAGUGGAAGUG------GAGUUGGCAGUGGG-GAGCCGGAGCAGCAUCACCUGGCUCCACUUGAGUU-------------AA------------A .((((((((.((..((...........(((------..((((.(..(((.-...)))..)))))..)))))..))))))))))...-------------..------------. ( -22.80) >DroYak_CAF1 147455 106 - 1 AUCGAGUGGGAGGAAGUGGAAGUGGAAGUGGAUUU------UGCAGCGGGCAAGUUGGAGCAGCAUCACCUGGCUCCACUUGAAUACGAACAAAG-CU-GAAAAAAAGCUGAAA .((((((((.((..((..((.(..(((.....)))------..).((..((........)).)).))..))..))))))))))..........((-((-.......)))).... ( -27.00) >consensus AUCGAGUGGGAGGAAGUGGAAGUGGAAGUGGAUUU__AGUUGGCAGUGGG_GAGCCGGAGCAGCAUCACCUGGCUCCACUUGAGUACGAA_AAA___U_GAAA_AAAG___AAA .((((((((............((...................))........((((((.(........)))))))))))))))............................... (-16.00 = -15.34 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:08 2006