
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
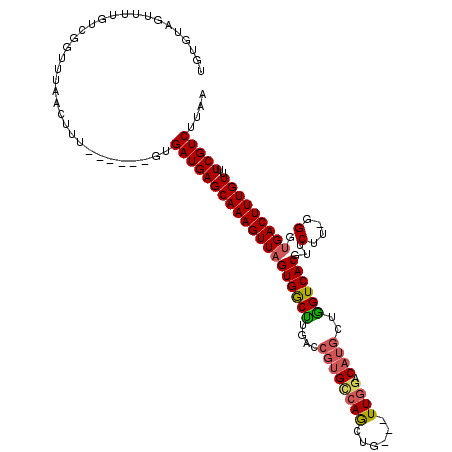
| Location | 21,678,698 – 21,678,806 |
| Length | 108 |
| Max. P | 0.559745 |

| Location | 21,678,698 – 21,678,806 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -22.26 |
| Energy contribution | -23.60 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
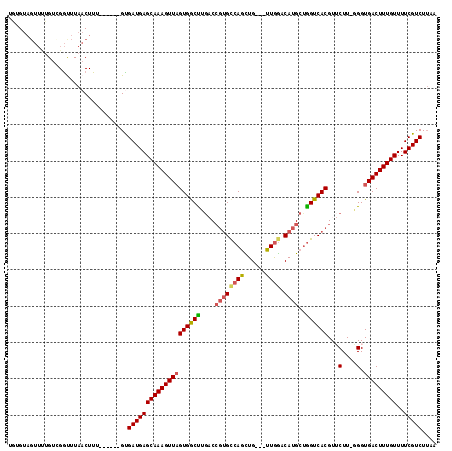
>3L_DroMel_CAF1 21678698 108 + 23771897 UGUGUAGUUUUGUCGGUUUAACUUU------GUGAUGAGCAAAGUUAGUGGCUUGACCGUGCCAGCUG---UUGGCCAUGCUGGUCACGUUCUUCGGGUGACUUUGUUUUCGUCU-AA .........................------..((((((((((((((.(.(...((.(((((((((((---.....)).))))).)))).))..).).)))))))))..))))).-.. ( -32.10) >DroPse_CAF1 21849 108 + 1 UAUGUAGUUUUGUUGGCUUAACUUU------GCGAUGAGCAAAGUUAGUGGCUUGACCGUGCCAGCUG---UUGGACAUGCUGGUCACGUUCUU-GGGUGACUUUGUUUUCGUCUUAA ..............(((.(((((((------((.....)))))))))((.(((..(.(((((((((((---.....)).))))).))))...).-.))).)).........))).... ( -35.00) >DroGri_CAF1 12413 110 + 1 ---G-----CAGCCGAUUUAACUUUGUGUUGGUGAUGAGCAAAGUUUGUGGCUGACCAGUGCCAGUUGUUGUUGACCAUGCUGGUCACGUACUUUUGGUGACUUUGUUUUCGUCUUAA ---.-----..((((((..........))))))(((((((((((((..(((....)))..(((((..(((((.((((.....))))))).))..)))))))))))))..))))).... ( -32.80) >DroWil_CAF1 45166 107 + 1 UGUGUAGUU-UGUCGGUUUAACUUU------GUGAUGAGCAAAGUUAGUGGCCAGCCCCGGUCAAUUG---UUAGACAUGCUGGUCACGUACUU-AGGUGACUUUGUUUUCGUCUUAA .........-.(.(((..(((((((------((.....)))))))))(((((((((....(((.....---...)))..)))))))))..((..-((....))..))..))).).... ( -33.00) >DroAna_CAF1 12898 109 + 1 UGUGUAGUUUUGUAGGUUUAACUUU------GUGAUGAGCAAAGUUAGUGACUUGACCGUGCCAGCUG---UUGGACAUGCAGGUCACGUUCUUCGGGUGACUUUGUUUUCGUCUUAA .........................------..((((((((((((((((((((((..((((((((...---)))).))))))))))))..((....)))))))))))..))))).... ( -32.00) >DroPer_CAF1 22026 108 + 1 UAUGUAGUUUUGUUGGCUUAACUUU------GCGAUGAGCAAAGUUAGUGGCUUGACCGUGCCAGCUG---UUGGACAUGCUAGUCACGUUCUU-GGGUGACUUUGUUUUCGUCUUAA ...........((((((((((((((------((.....)))))))))).((.....))..))))))..---..((((..((.((((((.(....-).))))))..))....))))... ( -32.60) >consensus UGUGUAGUUUUGUCGGUUUAACUUU______GUGAUGAGCAAAGUUAGUGGCUUGACCGUGCCAGCUG___UUGGACAUGCUGGUCACGUUCUU_GGGUGACUUUGUUUUCGUCUUAA .................................((((((((((((((((((((....((((((((......)))).))))..))))))...(....).)))))))))..))))).... (-22.26 = -23.60 + 1.34)

| Location | 21,678,698 – 21,678,806 |
|---|---|
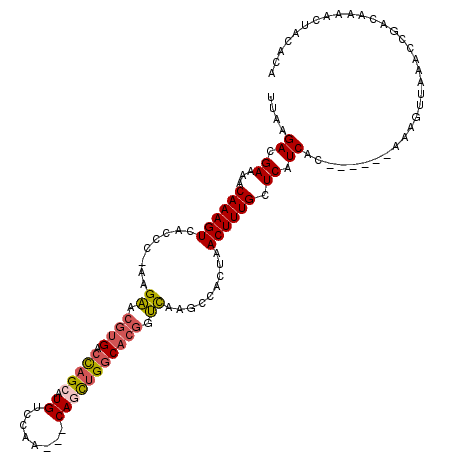
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -13.49 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
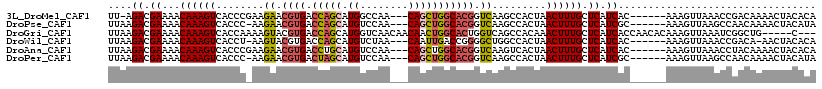
>3L_DroMel_CAF1 21678698 108 - 23771897 UU-AGACGAAAACAAAGUCACCCGAAGAACGUGACCAGCAUGGCCAA---CAGCUGGCACGGUCAAGCCACUAACUUUGCUCAUCAC------AAAGUUAAACCGACAAAACUACACA ..-.............(((.......((.((((.(((((.((.....---))))))))))).)).......((((((((.......)------)))))))....)))........... ( -27.50) >DroPse_CAF1 21849 108 - 1 UUAAGACGAAAACAAAGUCACCC-AAGAACGUGACCAGCAUGUCCAA---CAGCUGGCACGGUCAAGCCACUAACUUUGCUCAUCGC------AAAGUUAAGCCAACAAAACUACAUA ....(((.........)))....-..((.((((.(((((.((.....---))))))))))).))..((...(((((((((.....))------))))))).))............... ( -30.00) >DroGri_CAF1 12413 110 - 1 UUAAGACGAAAACAAAGUCACCAAAAGUACGUGACCAGCAUGGUCAACAACAACUGGCACUGGUCAGCCACAAACUUUGCUCAUCACCAACACAAAGUUAAAUCGGCUG-----C--- ....(((.........))).............((((((....((((........)))).))))))((((...(((((((.............))))))).....)))).-----.--- ( -23.62) >DroWil_CAF1 45166 107 - 1 UUAAGACGAAAACAAAGUCACCU-AAGUACGUGACCAGCAUGUCUAA---CAAUUGACCGGGGCUGGCCACUAACUUUGCUCAUCAC------AAAGUUAAACCGACA-AACUACACA ................(((....-......(((.(((((..(((...---.....)))....))))).)))((((((((.......)------)))))))....))).-......... ( -25.30) >DroAna_CAF1 12898 109 - 1 UUAAGACGAAAACAAAGUCACCCGAAGAACGUGACCUGCAUGUCCAA---CAGCUGGCACGGUCAAGUCACUAACUUUGCUCAUCAC------AAAGUUAAACCUACAAAACUACACA ....(((.........))).......((...((((((((.((.....---))....))).)))))..))..((((((((.......)------))))))).................. ( -21.50) >DroPer_CAF1 22026 108 - 1 UUAAGACGAAAACAAAGUCACCC-AAGAACGUGACUAGCAUGUCCAA---CAGCUGGCACGGUCAAGCCACUAACUUUGCUCAUCGC------AAAGUUAAGCCAACAAAACUACAUA ....(((.........)))....-..((.((((.(((((.((.....---))))))))))).))..((...(((((((((.....))------))))))).))............... ( -27.70) >consensus UUAAGACGAAAACAAAGUCACCC_AAGAACGUGACCAGCAUGUCCAA___CAGCUGGCACGGUCAAGCCACUAACUUUGCUCAUCAC______AAAGUUAAACCGACAAAACUACACA ....((.((...((((((........((.((((.(((((.((........))))))))))).)).........)))))).)).))................................. (-13.49 = -13.80 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:36 2006