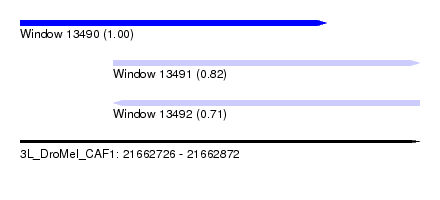
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,662,726 – 21,662,872 |
| Length | 146 |
| Max. P | 0.995602 |

| Location | 21,662,726 – 21,662,838 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -17.61 |
| Energy contribution | -19.43 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21662726 112 + 23771897 ACCA------CUUUGCUCACUGAAAUCGCAGACGUGUGGCAACACUC-AAGCCGCCAGAAAGAUCUGGCAUCUCUCAUACUUA-ACACGCGCAUGCAAAAUAUUUAAGAAAUAAAUAUUA ....------.................(((..((((((((.......-..)))((((((....))))))..............-..)))))..)))..((((((((.....)))))))). ( -27.50) >DroEre_CAF1 39024 114 + 1 ACCA------CUUUACGCACUGAAACAGUAGUGGUGUGGCAACGCUCGAAGCCGCCAGAAAGAUCUGGCAUCGCCGAAUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGGAAAUUA ((((------((......((((...))))))))))(((....)))........((((((....))))))....(((..(((((((...((....))......)))))))..)))...... ( -31.90) >DroYak_CAF1 48015 114 + 1 ACCA------CUUUGCACACUGAAACCGCAGUGGUGUGGCAACGCCCGAAGCCGCCAGAAAGGUCUGGCAACGCUGAUUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGGAAAUCA .(((------.......(((((......)))))(((((((..(....)..)))((((((....)))))).))))...((((((((...((....))......)))))))).)))...... ( -34.40) >DroAna_CAF1 38727 117 + 1 ACCCUUAUCUUUUCCAACACUGUCAUUGCAAAGAUAUGGCAGCACC-AAAGCCGCGAAAGAGAUAUGGCAGCAUUGAUUUAGAAACACGCGCAAGCUAAAUAUUAAACAAAUGAGAAA-- ..((.((((((((.(..((.((((........))))))((.((...-...)).))).)))))))).))...((((..(((((......((....))......)))))..)))).....-- ( -22.20) >consensus ACCA______CUUUGCACACUGAAACCGCAGAGGUGUGGCAACACCCGAAGCCGCCAGAAAGAUCUGGCAUCGCUGAUUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGAAAAUUA .................(((((......)))))(((((((..........)))((((((....))))))...............))))((....))........................ (-17.61 = -19.43 + 1.81)

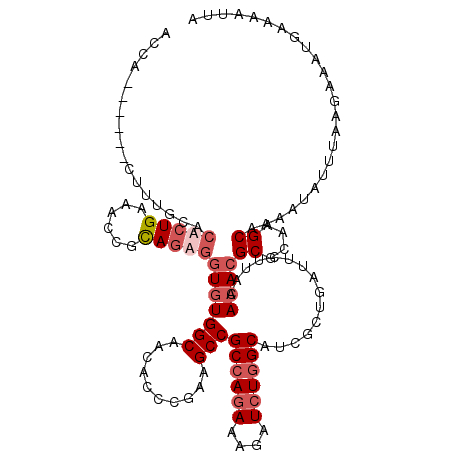
| Location | 21,662,760 – 21,662,872 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.82 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21662760 112 + 23771897 AACACUC-AAGCCGCCAGAAAGAUCUGGCAUCUCUCAUACUUA-ACACGCGCAUGCAAAAUAUUUAAGAAAUAAAUAUUAGUCGCUAAC---AGUUUAAAUAUAAUAAAAUUCUGCU .......-.(((.((((((....))))))..............-....(((.((....((((((((.....)))))))).)))))....---......................))) ( -17.40) >DroEre_CAF1 39058 117 + 1 AACGCUCGAAGCCGCCAGAAAGAUCUGGCAUCGCCGAAUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGGAAAUUAGUAGCUAACAAUAGUUUAAAUAUUAUAAUAUUCUGCU ...((.....)).((((((....))))))...((.((((.(((.....((....))..(((((((((..(((....)))....((((....))))))))))))).))).)))).)). ( -26.10) >DroYak_CAF1 48049 117 + 1 AACGCCCGAAGCCGCCAGAAAGGUCUGGCAACGCUGAUUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGGAAAUCAGUAGCUAACAAUAGUGUAAAUAUUAUAAUAUUCUGCU ...((....(((.((((((....))))))...(((((((((((((...((....))......))))))).......)))))).)))....((((((....))))))........)). ( -26.11) >DroAna_CAF1 38767 105 + 1 AGCACC-AAAGCCGCGAAAGAGAUAUGGCAGCAUUGAUUUAGAAACACGCGCAAGCUAAAUAUUAAACAAAUGAGAAA---UAGCUAAU--------AAAUACUAUUUAAAACGCUU (((...-...(((((....).....))))..((((..(((((......((....))......)))))..))))..(((---(((.....--------.....)))))).....))). ( -15.90) >consensus AACACCCGAAGCCGCCAGAAAGAUCUGGCAUCGCUGAUUCUUAAACACGCGCAAGCAAAAUAUUUAAGAAAUGAAAAUUAGUAGCUAAC___AGUUUAAAUAUUAUAAAAUUCUGCU .........(((.((((((....))))))...................((....))...........................)))............................... (-13.50 = -14.00 + 0.50)

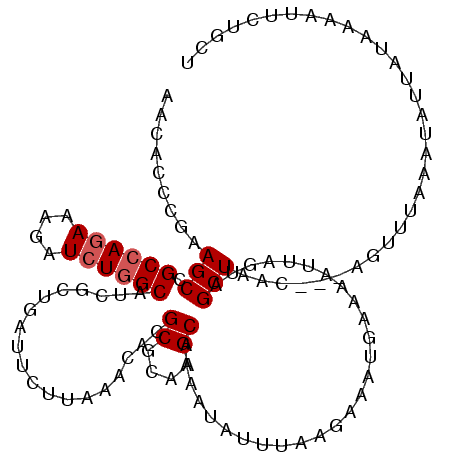
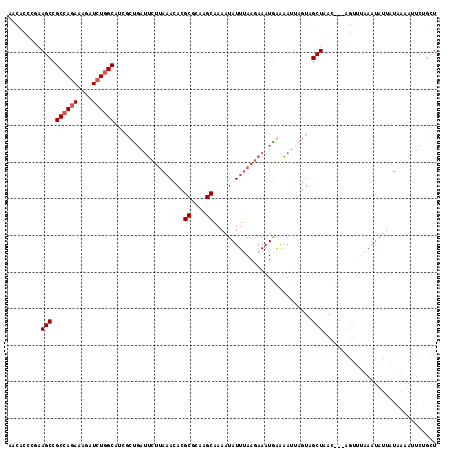
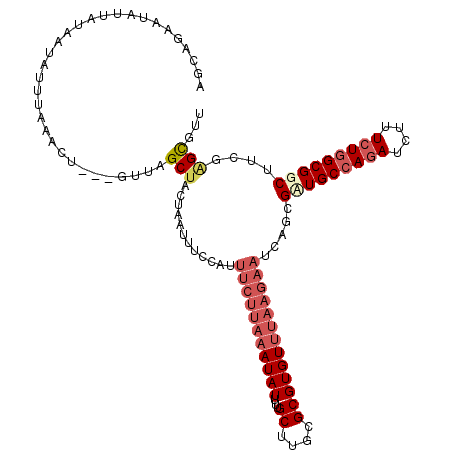
| Location | 21,662,760 – 21,662,872 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -15.04 |
| Energy contribution | -17.60 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21662760 112 - 23771897 AGCAGAAUUUUAUUAUAUUUAAACU---GUUAGCGACUAAUAUUUAUUUCUUAAAUAUUUUGCAUGCGCGUGU-UAAGUAUGAGAGAUGCCAGAUCUUUCUGGCGGCUU-GAGUGUU (((...(((((.(((((((((((((---((..((((..((((((((.....))))))))))))..))).)).)-))))))))))))))((((((....)))))).))).-....... ( -28.70) >DroEre_CAF1 39058 117 - 1 AGCAGAAUAUUAUAAUAUUUAAACUAUUGUUAGCUACUAAUUUCCAUUUCUUAAAUAUUUUGCUUGCGCGUGUUUAAGAUUCGGCGAUGCCAGAUCUUUCUGGCGGCUUCGAGCGUU .((.((((((....))))))......(((..((((....(((.((...((((((((((...((....))))))))))))...)).)))((((((....)))))))))).)))))... ( -29.70) >DroYak_CAF1 48049 117 - 1 AGCAGAAUAUUAUAAUAUUUACACUAUUGUUAGCUACUGAUUUCCAUUUCUUAAAUAUUUUGCUUGCGCGUGUUUAAGAAUCAGCGUUGCCAGACCUUUCUGGCGGCUUCGGGCGUU .((.((((((....))))))................((((.......(((((((((((...((....))))))))))))).....(((((((((....))))))))).))))))... ( -29.50) >DroAna_CAF1 38767 105 - 1 AAGCGUUUUAAAUAGUAUUU--------AUUAGCUA---UUUCUCAUUUGUUUAAUAUUUAGCUUGCGCGUGUUUCUAAAUCAAUGCUGCCAUAUCUCUUUCGCGGCUUU-GGUGCU ..((((...(((((((....--------....))))---))).......(((........)))..))))..........(((((.(((((.(........).))))).))-)))... ( -18.40) >consensus AGCAGAAUAUUAUAAUAUUUAAACU___GUUAGCUACUAAUUUCCAUUUCUUAAAUAUUUUGCUUGCGCGUGUUUAAGAAUCAGCGAUGCCAGAUCUUUCUGGCGGCUUCGAGCGUU ................................(((............(((((((((((...((....))))))))))))).....(((((((((....)))))))))....)))... (-15.04 = -17.60 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:29 2006