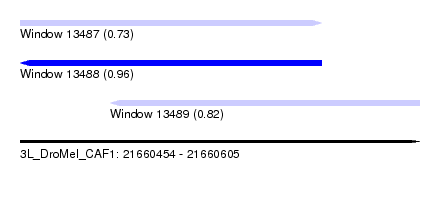
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,660,454 – 21,660,605 |
| Length | 151 |
| Max. P | 0.964731 |

| Location | 21,660,454 – 21,660,568 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -48.95 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
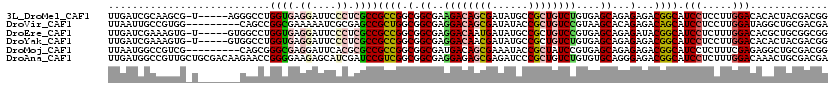
>3L_DroMel_CAF1 21660454 114 + 23771897 UUGAUCGCAAGCG-U-----AGGGCCUGGUGAGGAUUCCCUCGCCGCCGGCGGCGAAGACAGCGAUAUGCCGCUGUCUGUGAGCAGAGAGACGGCAUCCUCCUUGGACACACUACGACGG .....((....((-(-----((.(((.((.((((((.((.((((((....))))))((((((((......))))))))..............)).)))))))).)).)...))))).)). ( -46.90) >DroVir_CAF1 22823 111 + 1 UUAAUUGCCGUGG---------CAGCCGGCGAAAAAUCGCGAGCCGCUGGCGGCGAGGACAGCGAUAUACCGCUGUCCGUAAGCACAGAGACAGCAUCCUCCUUGGAUAGGCUGCGACGA ........(((.(---------(((((.((((....)))).....((((.(.((..((((((((......))))))))....))...)...))))((((.....)))).)))))).))). ( -51.70) >DroEre_CAF1 36725 114 + 1 UUGAUCGAAAGUG-U-----GUGGCCUGGUGAGGAUUCCCUCGCCGCCGGCGGCGAGGACAAUGAUAUGCCGCUGUCCGUGAGCAGAGAUACGGCAUCCUCUUUGGACACGCUGCGGCGG .....((...(((-(-----(((..(..(.(((((...((((((((....)))))))).........(((((.(((((....)....)))))))))))))).)..).))))).))..)). ( -48.10) >DroYak_CAF1 45744 114 + 1 UUGAUCGAAAGUG-U-----GUGGCCUGGUGAGGAUUCCCUCGCCGCCGGCGGCGAGGACAACGAUAUGCCGCUGUCUGUGAGCAGAGAGACGGCAUCCUCCUUGGACACACUACGACGG ....(((..((((-(-----((..((.((.(((((...((((((((....)))))))).........(((((((.((((....)))).)).)))))))))))).))))))))).)))... ( -55.70) >DroMoj_CAF1 25475 111 + 1 UUAAUGGCCGUCG---------CAGCGGGCGAGGAUUCACGCGCCGCCGGCGGCGAUGACAGCGAAAUACCGCUAUCCGUGAGCAGAGAGACGGCAUCCUCUUUCGAGAGGCUGCGACGG .......((((((---------((((....((((((((((((((((....)))))..((.((((......)))).)))))))((.(.....).))))))))...(....))))))))))) ( -50.30) >DroAna_CAF1 36287 120 + 1 UUGAUGGCCGUUGCUGCGACAAGAACCGGGGAAGAGCAUCGAUCCGUCGGCGGCGAGGAGAGCGAGAUCCCGCUGUCUGUGUGCAGGGAGACGGCAUCCUCUUUGGACAAACUGCGACGA ((((((..(....((.((........)).))..)..))))))..(((((((((((.(((........))))))))))..(((.(((((((........))))))).)))......)))). ( -41.00) >consensus UUGAUCGCAAGUG_U_______GACCUGGUGAGGAUUCCCGCGCCGCCGGCGGCGAGGACAGCGAUAUGCCGCUGUCCGUGAGCAGAGAGACGGCAUCCUCCUUGGACACACUGCGACGG ...........................((((.((....)).))))((((.(.((..((((((((......))))))))....))...)...)))).(((.....)))............. (-26.34 = -26.98 + 0.64)


| Location | 21,660,454 – 21,660,568 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -47.12 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
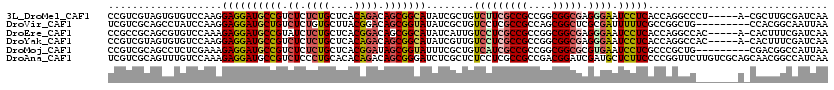
>3L_DroMel_CAF1 21660454 114 - 23771897 CCGUCGUAGUGUGUCCAAGGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGCUGUCUUCGCCGCCGGCGGCGAGGGAAUCCUCACCAGGCCCU-----A-CGCUUGCGAUCAA ..(((((((((((.((...((((((((((.((.((((....)))).)))))).........(((((((((....))))))))).))))))....))...)-----)-))).))))))... ( -48.80) >DroVir_CAF1 22823 111 - 1 UCGUCGCAGCCUAUCCAAGGAGGAUGCUGUCUCUGUGCUUACGGACAGCGGUAUAUCGCUGUCCUCGCCGCCAGCGGCUCGCGAUUUUUCGCCGGCUG---------CCACGGCAAUUAA ..(..(((((..((((.....)))))))))..).........(((((((((....)))))))))..((((...((((((.((((....)))).)))))---------)..))))...... ( -49.30) >DroEre_CAF1 36725 114 - 1 CCGCCGCAGCGUGUCCAAAGAGGAUGCCGUAUCUCUGCUCACGGACAGCGGCAUAUCAUUGUCCUCGCCGCCGGCGGCGAGGGAAUCCUCACCAGGCCAC-----A-CACUUUCGAUCAA ..((....))(((.((...(((((((((((...((((....))))..))))).........(((((((((....))))))))).))))))....)).)))-----.-............. ( -43.60) >DroYak_CAF1 45744 114 - 1 CCGUCGUAGUGUGUCCAAGGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGUUGUCCUCGCCGCCGGCGGCGAGGGAAUCCUCACCAGGCCAC-----A-CACUUUCGAUCAA ..((((.(((((((((...((((((((((.((.((((....)))).)))))).........(((((((((....))))))))).))))))....))..))-----)-))))..))))... ( -53.60) >DroMoj_CAF1 25475 111 - 1 CCGUCGCAGCCUCUCGAAAGAGGAUGCCGUCUCUCUGCUCACGGAUAGCGGUAUUUCGCUGUCAUCGCCGCCGGCGGCGCGUGAAUCCUCGCCCGCUG---------CGACGGCCAUUAA ((((((((((.........((((((((.(.....).))(((((((((((((....))))))))..(((((....))))))))))))))))....))))---------))))))....... ( -53.82) >DroAna_CAF1 36287 120 - 1 UCGUCGCAGUUUGUCCAAAGAGGAUGCCGUCUCCCUGCACACAGACAGCGGGAUCUCGCUCUCCUCGCCGCCGACGGAUCGAUGCUCUUCCCCGGUUCUUGUCGCAGCAACGGCCAUCAA ............((((.....))))(((((....(((((((..(((.(.((((....((((((((((....))).)))..)).))...))))).)))..))).))))..)))))...... ( -33.60) >consensus CCGUCGCAGCGUGUCCAAAGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGCUGUCCUCGCCGCCGGCGGCGAGGGAAUCCUCACCAGGCC_______A_CAACGGCGAUCAA ...................((((((((((.((.((((....)))).)))))))........(((((((((....))))).)))).))))).............................. (-24.33 = -24.72 + 0.39)


| Location | 21,660,488 – 21,660,605 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -25.91 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
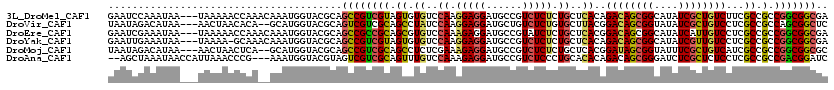
>3L_DroMel_CAF1 21660488 117 - 23771897 GAAUCCAAAUAA---UAAAAACCAAACAAAUGGUACGCAGCCGUCGUAGUGUGUCCAAGGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGCUGUCUUCGCCGCCGGCGGCGA ............---.....((((......)))).(((.((((.((...((((..((..((((......))))..))..))))((((((((....)))))))).....)).)))).))). ( -39.90) >DroVir_CAF1 22854 115 - 1 UAAUAGACAUAA---AACUAACACA--GCAUGGUACGCAGUCGUCGCAGCCUAUCCAAGGAGGAUGCUGUCUCUGUGCUUACGGACAGCGGUAUAUCGCUGUCCUCGCCGCCAGCGGCUC ............---.........(--((((((...((((.((((....(((.....)))..))))))))..)))))))...(((((((((....)))))))))..((((....)))).. ( -41.30) >DroEre_CAF1 36759 117 - 1 GAAUCGAAAUAA---UAAAAACCAAACAAAUGGUACGCAGCCGCCGCAGCGUGUCCAAAGAGGAUGCCGUAUCUCUGCUCACGGACAGCGGCAUAUCAUUGUCCUCGCCGCCGGCGGCGA ............---.....((((......)))).....((((((((.(((.(..(((.((..(((((((...((((....))))..))))))).)).)))..).))).).))))))).. ( -44.30) >DroYak_CAF1 45778 116 - 1 GAAUUGAAAUAA---UAAAA-GCAAACAAAUGGUACGCAGCCGUCGUAGUGUGUCCAAGGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGUUGUCCUCGCCGCCGGCGGCGA ............---.....-((..((.....))..)).(((((((..(((.(..(((.((..((((((.((.((((....)))).)))))))).)).)))..).)))..).)))))).. ( -40.50) >DroMoj_CAF1 25506 115 - 1 UAAUAGACAUAA---AACUAACUCA--GCAUGGUACGCAGCCGUCGCAGCCUCUCGAAAGAGGAUGCCGUCUCUCUGCUCACGGAUAGCGGUAUUUCGCUGUCAUCGCCGCCGGCGGCGC .....(((....---..........--(((((((.....))))).))(((..(((....)))(((((((.((.((((....)))).)))))))))..))))))..(((((....))))). ( -40.50) >DroAna_CAF1 36327 115 - 1 --AGCUAAAUAACCAUUAAACCCG---AAAUGGUACGUAGUCGUCGCAGUUUGUCCAAAGAGGAUGCCGUCUCCCUGCACACAGACAGCGGGAUCUCGCUCUCCUCGCCGCCGACGGAUC --.((((....((((((.......---.))))))...))))((((((.((((((....((.(((.......)))))....)))))).((((((........))).))).).))))).... ( -31.20) >consensus GAAUCGAAAUAA___UAAAAACCAA__AAAUGGUACGCAGCCGUCGCAGCGUGUCCAAAGAGGAUGCCGUCUCUCUGCUCACAGACAGCGGCAUAUCGCUGUCCUCGCCGCCGGCGGCGA .......................................((((((((.((.((..((.(((((......))))).))..))..((((((((....))))))))...)).).))))))).. (-25.91 = -25.55 + -0.36)
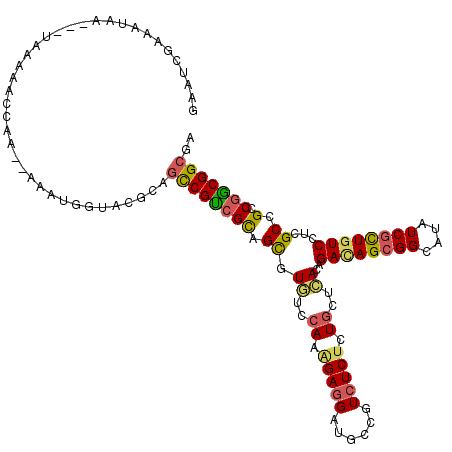

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:27 2006