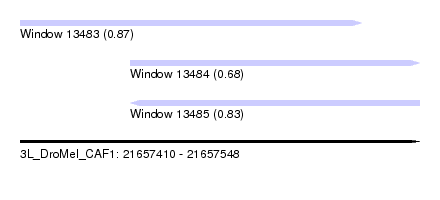
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,657,410 – 21,657,548 |
| Length | 138 |
| Max. P | 0.871225 |

| Location | 21,657,410 – 21,657,528 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -36.31 |
| Consensus MFE | -22.05 |
| Energy contribution | -23.30 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21657410 118 + 23771897 CCACAGCGCCUGCAGAUGCCACCGCUGCUCAGAG-AGCUCGGUUCGCAUGCAGUGCGGGGCUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGC ((...((((.((((..(((.((((..((((...)-))).))))..))))))))))).))(((((.((((((...(((.((((((..........)))))).)))..))))))..))))) ( -42.10) >DroEre_CAF1 33648 116 + 1 GCACAACGCCUGCAGAUGCCACCGCUGCUCAG---AGCUCAGUUCGCAUGCAGUGCGGUAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGC ((((((.....(((((((((..((((((...(---(((...))))....)))))).))))))))).........(((.((((((..........)))))).)))...))))))...... ( -41.20) >DroYak_CAF1 42786 116 + 1 CCACAACGCUUGCAGAUGCCACCGCUGCUCAG---AGCUCAGUUCGCUUCCAGUGCGGUAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGC .......(((((((((((((..(((((....(---(((.......)))).))))).)))))))))((((((...(((.((((((..........)))))).)))..))))))...)))) ( -34.50) >DroAna_CAF1 33800 104 + 1 ------------UGGAUCCCA--ACUGCUUGGCGACGAUCGGUUCGCAUGCAGUGUGCCGUUUGCCAUAAAA-AUAAUUGCCGCCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAAC ------------.((......--(((((...((((........))))..)))))...))(((((.((((((.-......(((((..........))))).......))))))..))))) ( -27.44) >consensus CCACAACGCCUGCAGAUGCCACCGCUGCUCAG___AGCUCAGUUCGCAUGCAGUGCGGUAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGC ...........(((((((((..((((((.....................)))))).)))))))))((((((...(((.((((((..........)))))).)))..))))))....... (-22.05 = -23.30 + 1.25)


| Location | 21,657,448 – 21,657,548 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21657448 100 + 23771897 CGGUUCGCAUGCAGUGCGGGGCUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGCCCAGACCAUUUAUGGCCAAC-------------------- .(((((((((...)))))(((((((.((((((...(((.((((((..........)))))).)))..))))))..))))))).)))).............-------------------- ( -34.10) >DroVir_CAF1 20138 83 + 1 --GU----------UGUACCA-UGGCCAUAAA-AAUAUUUGCCGCCUUAAUGUGAGUGGCUUAUAAAUUUG--CACAAACGC-GACCAUAUAUAGCAUUU-------------------- --((----------((((..(-(((.......-..(((..((((((.......).)))))..)))...(((--(......))-)))))).))))))....-------------------- ( -19.40) >DroEre_CAF1 33684 100 + 1 CAGUUCGCAUGCAGUGCGGUAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGCCCAGGCCAUUUAUGGCCAUC-------------------- ......((((...))))(((.(.(((.(((((...(((.((((((..........)))))).)))..)))))))).).)))..(((((....)))))...-------------------- ( -27.50) >DroYak_CAF1 42822 100 + 1 CAGUUCGCUUCCAGUGCGGUAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAGCCCAAGCCAUUUAUGGCCAUC-------------------- ......((((...(((((((....)))(((((...(((.((((((..........)))))).)))..)))))))))))))....((((....))))....-------------------- ( -25.10) >DroMoj_CAF1 22976 83 + 1 --GU----------UGCGCCA-UGGCCAUAAA-AAUAUUUGCCGCCUUAAUGUGAGUGGCUUAUAAAUUUG--CACAAACGC-GACCAUAUAUAGCGGCU-------------------- --((----------(((...(-(((.......-..(((..((((((.......).)))))..)))...(((--(......))-)))))).....))))).-------------------- ( -20.20) >DroAna_CAF1 33825 119 + 1 CGGUUCGCAUGCAGUGUGCCGUUUGCCAUAAAA-AUAAUUGCCGCCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAACCCAGACCAUUUAUGGCCAUGUGGCGGAGCCAGGGCGAGGG ((((.(((.....))).))))((((((......-......(((((.....(((((((((((......((((....))))...)).))))))))).....)))))(....)..)))))).. ( -31.90) >consensus C_GUUCGCAUGCAGUGCGCCAUUUGCCAUAAAAAAUAUUGGCCACCUUAAUGUAAGUGGCUUAUAAAUUUGUGCACAAACCCAGACCAUUUAUGGCCAUC____________________ ........................((((((((...(((..(((((.(((...))))))))..)))..((((....)))).........))))))))........................ (-14.17 = -14.28 + 0.11)



| Location | 21,657,448 – 21,657,548 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21657448 100 - 23771897 --------------------GUUGGCCAUAAAUGGUCUGGGCUUGUGCACAAAUUUAUAAGCCACUUACAUUAAGGUGGCCAAUAUUUUUUAUGGCAAGCCCCGCACUGCAUGCGAACCG --------------------...(((((....))))).((((((((.((.(((..(((..(((((((......)))))))..)))...))).))))))))))((((.....))))..... ( -34.40) >DroVir_CAF1 20138 83 - 1 --------------------AAAUGCUAUAUAUGGUC-GCGUUUGUG--CAAAUUUAUAAGCCACUCACAUUAAGGCGGCAAAUAUU-UUUAUGGCCA-UGGUACA----------AC-- --------------------..........(((((((-(((....))--)(((..(((..(((.((........)).)))..)))..-)))..)))))-)).....----------..-- ( -16.70) >DroEre_CAF1 33684 100 - 1 --------------------GAUGGCCAUAAAUGGCCUGGGCUUGUGCACAAAUUUAUAAGCCACUUACAUUAAGGUGGCCAAUAUUUUUUAUGGCAAAUACCGCACUGCAUGCGAACUG --------------------....(((((((((((((..((((((((........))))))))((((......)))))))))......))))))))......((((.....))))..... ( -31.70) >DroYak_CAF1 42822 100 - 1 --------------------GAUGGCCAUAAAUGGCUUGGGCUUGUGCACAAAUUUAUAAGCCACUUACAUUAAGGUGGCCAAUAUUUUUUAUGGCAAAUACCGCACUGGAAGCGAACUG --------------------....(((((((((((((..((((((((........))))))))((((......)))))))))......))))))))......(((.......)))..... ( -28.10) >DroMoj_CAF1 22976 83 - 1 --------------------AGCCGCUAUAUAUGGUC-GCGUUUGUG--CAAAUUUAUAAGCCACUCACAUUAAGGCGGCAAAUAUU-UUUAUGGCCA-UGGCGCA----------AC-- --------------------.((.(((....((((((-(((....))--)(((..(((..(((.((........)).)))..)))..-)))..)))))-)))))).----------..-- ( -19.90) >DroAna_CAF1 33825 119 - 1 CCCUCGCCCUGGCUCCGCCACAUGGCCAUAAAUGGUCUGGGUUUGUGCACAAAUUUAUAAGCCACUUACAUUAAGGCGGCAAUUAU-UUUUAUGGCAAACGGCACACUGCAUGCGAACCG ...((((....((...(((.....((((((((.....((((((((....))))))))...(((.(((......))).)))......-.))))))))....))).....))..)))).... ( -31.70) >consensus ____________________GAUGGCCAUAAAUGGUCUGGGCUUGUGCACAAAUUUAUAAGCCACUUACAUUAAGGCGGCAAAUAUUUUUUAUGGCAAAUGCCGCACUGCAUGCGAAC_G ........................((((((((..(((..((((((((........)))))))).(((.....)))..)))........))))))))........................ (-14.31 = -14.48 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:23 2006