
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
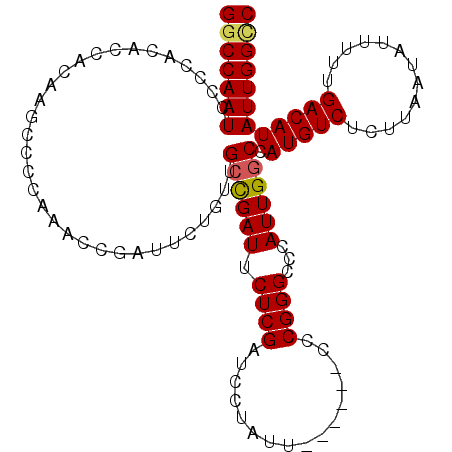
| Location | 21,654,171 – 21,654,275 |
| Length | 104 |
| Max. P | 0.882554 |

| Location | 21,654,171 – 21,654,275 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21654171 104 + 23771897 GGCCAAUCCCCAUACCACAAGCCCCAAAAUGAUUCUGUUGCUGAUUCUCGAUCCUAUU------CCCGGGCCCAUUGUCCAUGUCUCUUAAUAUUUUUGACAUAUUGGCC (((((((.........((((((((......((.((.......)).))..((......)------)..))))...))))..(((((.............)))))))))))) ( -20.82) >DroSec_CAF1 31155 104 + 1 GGCCAAUCCCCACACCACAAGCCCCAAACCGAUUCUGUUGCCGAUUCUCGAUCCUAUU------CCCGGGCCCAUUGGCCAUGUCUCUUAAUAUUUUUGACAUAUUGGCC (((((((.......(((...((((......((.((.......)).))..((......)------)..))))....)))..(((((.............)))))))))))) ( -22.02) >DroSim_CAF1 35607 104 + 1 GGCCAAUCCCCACACCACAAGCCCCAAACCGAUUCUGUUGCCGAUUCUCGAUCCUAUU------CCCGGGCCCAUUGGCCAUGUCUCUUAAUAUUUUUGACAUAUUGGCC (((((((.......(((...((((......((.((.......)).))..((......)------)..))))....)))..(((((.............)))))))))))) ( -22.02) >DroEre_CAF1 30476 104 + 1 GGCCAAUCCCCACACCACAAACCCAAAACCGAUUCUGUUGCCGAUUCUCGAUCCUAUU------CCCGGGCCCAUUGGCCAUGUCUCUUAAUAUUUUUGACAUAUUGGUC (((((((.(((...................((.((.......)).))..((......)------)..)))...)))))))(((((.............)))))....... ( -19.72) >DroYak_CAF1 39573 109 + 1 GGCCAAUCCCCACACCACAAACCCAAAACCGAUUCUGUUGCCGAUUCUCGAUCCUAUUCUCGAUCCCGGGCCCAUUGGCCAUGUCUCUUAAUAUUUUUGACAUAUUGG-C (((((((.(((...................((.((.......)).))..((((........))))..)))...)))))))(((((.............))))).....-. ( -22.82) >consensus GGCCAAUCCCCACACCACAAGCCCCAAACCGAUUCUGUUGCCGAUUCUCGAUCCUAUU______CCCGGGCCCAUUGGCCAUGUCUCUUAAUAUUUUUGACAUAUUGGCC (((((((................................((((((.((((................))))...)))))).(((((.............)))))))))))) (-17.53 = -17.65 + 0.12)


| Location | 21,654,171 – 21,654,275 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.86 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21654171 104 - 23771897 GGCCAAUAUGUCAAAAAUAUUAAGAGACAUGGACAAUGGGCCCGGG------AAUAGGAUCGAGAAUCAGCAACAGAAUCAUUUUGGGGCUUGUGGUAUGGGGAUUGGCC ((((((((((((.............)))))..((.(..(((((.((------(((.((.((..............)).))))))).)))))..).))......))))))) ( -29.96) >DroSec_CAF1 31155 104 - 1 GGCCAAUAUGUCAAAAAUAUUAAGAGACAUGGCCAAUGGGCCCGGG------AAUAGGAUCGAGAAUCGGCAACAGAAUCGGUUUGGGGCUUGUGGUGUGGGGAUUGGCC ((((((((((((.............))))).(((.(..(((((.(.------.....((((((...(((....).)).))))))).)))))..))))......))))))) ( -34.22) >DroSim_CAF1 35607 104 - 1 GGCCAAUAUGUCAAAAAUAUUAAGAGACAUGGCCAAUGGGCCCGGG------AAUAGGAUCGAGAAUCGGCAACAGAAUCGGUUUGGGGCUUGUGGUGUGGGGAUUGGCC ((((((((((((.............))))).(((.(..(((((.(.------.....((((((...(((....).)).))))))).)))))..))))......))))))) ( -34.22) >DroEre_CAF1 30476 104 - 1 GACCAAUAUGUCAAAAAUAUUAAGAGACAUGGCCAAUGGGCCCGGG------AAUAGGAUCGAGAAUCGGCAACAGAAUCGGUUUUGGGUUUGUGGUGUGGGGAUUGGCC .......(((((.............)))))(((((((...((((.(------....(((((.((((((((........)))))))).)))))....).)))).))))))) ( -30.32) >DroYak_CAF1 39573 109 - 1 G-CCAAUAUGUCAAAAAUAUUAAGAGACAUGGCCAAUGGGCCCGGGAUCGAGAAUAGGAUCGAGAAUCGGCAACAGAAUCGGUUUUGGGUUUGUGGUGUGGGGAUUGGCC (-((((((((((.............))))).((((.(..((((((((((((......(.(((.....)))).......))))))))))))..)))))......)))))). ( -36.04) >consensus GGCCAAUAUGUCAAAAAUAUUAAGAGACAUGGCCAAUGGGCCCGGG______AAUAGGAUCGAGAAUCGGCAACAGAAUCGGUUUGGGGCUUGUGGUGUGGGGAUUGGCC ((((((((((((.............))))).(((.(..(((((.............(((((((...(((....).)).))))))).)))))..))))......))))))) (-24.90 = -25.86 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:17 2006