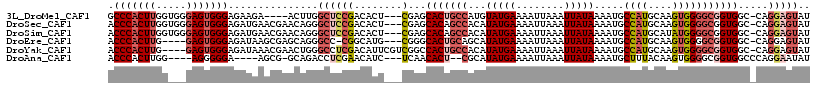
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
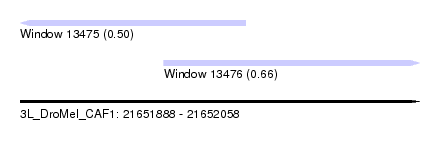
| Location | 21,651,888 – 21,652,058 |
| Length | 170 |
| Max. P | 0.655809 |

| Location | 21,651,888 – 21,651,984 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
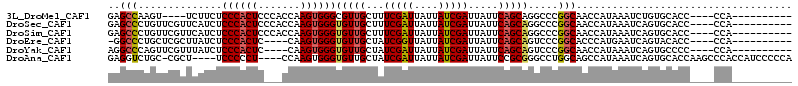
>3L_DroMel_CAF1 21651888 96 - 23771897 GAGCCAAGU----UCUUCUCCCACUCCCACCAAGUGGGCGUUGCUUUCGAUUAUUAUCGAUUAUUCAGCAGGCCCGGCAACCAUAAAUCUGUGCACC----CCA---------- (.(((....----......((((((.......)))))).((((...(((((....))))).....)))).))).)((....((((....))))....----)).---------- ( -23.00) >DroSec_CAF1 28624 100 - 1 GAGCCCUGUUCGUUCAUCUCCCACUCCCACCAAGUGGGUGUUGCUUUCGAUUAUUAUCGAUUAUUCAGCAGGCCCGGCAACCAUAAAUCAGUGCACC----CCA---------- (.(((..............((((((.......))))))(((((...(((((....))))).....)))))))).)((....(((......)))....----)).---------- ( -22.10) >DroSim_CAF1 33059 100 - 1 GAGCCCUGUUCGUUCAUCUCCCACUCCCACCAAGUGGGUGUUGCUUUCGAUUAUUAUCGAUUAUUCAGCAGGCCCGGCAACCAUAAAUCAGUGCACC----CCA---------- (.(((..............((((((.......))))))(((((...(((((....))))).....)))))))).)((....(((......)))....----)).---------- ( -22.10) >DroEre_CAF1 27700 95 - 1 -GGCCCUGCUCGCUUAUCUCCCACUC----CAAGUGGGUGUUGCUAUCGGUUAUUAUCGAUUAUUCAGCAGUCCCGGCACCCAUGAAUCAGUACACC----CCA---------- -((..(((((.((....(.(((((..----...))))).)..)).((((((....)))))).....)))))..))......................----...---------- ( -24.40) >DroYak_CAF1 36919 96 - 1 AGGCCCAGUUCGUUUAUCUCCCACUC----CAAGUGGGUGUUGCUAUCGAUUAUUAUCGAUUAUUCAGCAGUCCCGGCAACCAUAAAUCAGUGCCCC----CCA---------- .((..(.............(((((..----...)))))(((((..((((((....))))))....))))))..))((((............))))..----...---------- ( -23.10) >DroAna_CAF1 25937 105 - 1 GAGGUCUGC-CGCU----UCCCCCU----CCAAGUGGGUGUUGCUAUCGAUUAUUAUCGAUUAUUCCGCGGGCCUGGCAGCCAUAAAUCAGUGCACCAAGCCCACCAUCCCCCA ..(((...(-((((----(......----..))))))(((..(..((((((....))))))..)..)))((((.(((..(((........).)).))).)))))))........ ( -30.00) >consensus GAGCCCUGUUCGUUCAUCUCCCACUC___CCAAGUGGGUGUUGCUAUCGAUUAUUAUCGAUUAUUCAGCAGGCCCGGCAACCAUAAAUCAGUGCACC____CCA__________ ..(((..............((((((.......))))))(((((...(((((....))))).....))))).....))).................................... (-17.25 = -17.53 + 0.28)
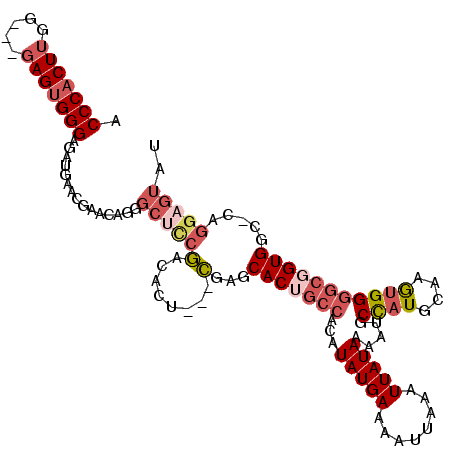
| Location | 21,651,949 – 21,652,058 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -20.86 |
| Energy contribution | -22.53 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
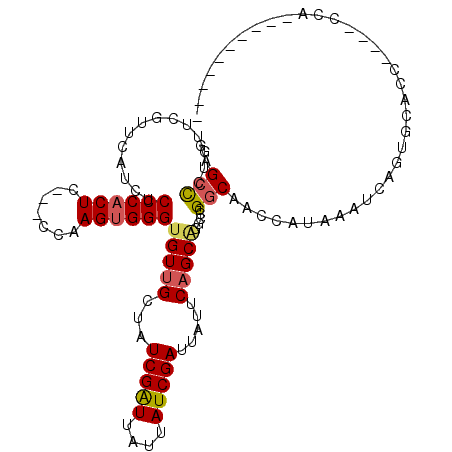
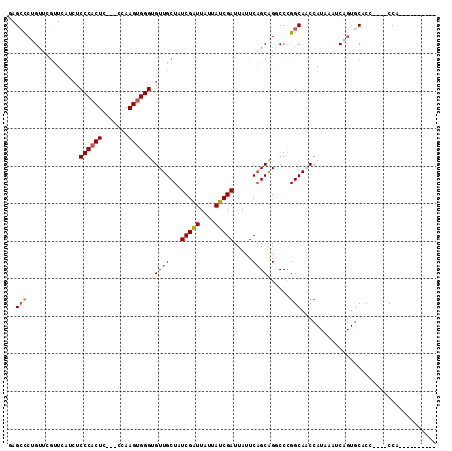
>3L_DroMel_CAF1 21651949 109 + 23771897 GCCCACUUGGUGGGAGUGGGAGAAGA----ACUUGGCUCCGACACU---CGAGCACUGCCAUGUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAAGUGGGGCGGUGGC-CAGGAGUAU .(((((((.....)))))))......----.((((((..((.....---))..(((((((...(((((........))))).....((((....)))))))))))))-))))..... ( -36.80) >DroSec_CAF1 28685 113 + 1 ACCCACUUGGUGGGAGUGGGAGAUGAACGAACAGGGCUCCGACACU---CGAGCACAGCCACAUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAAGUGGGGCGGUGGC-CAGGAGUAU .....(((((((((.((.((((.((......))...)))).)).))---)...(((.(((...(((((........))))).....((((....))))))).)))))-))))..... ( -33.40) >DroSim_CAF1 33120 113 + 1 ACCCACUUGGUGGGAGUGGGAGAUGAACGAACAGGGCUCCGACACU---CGAGCACAGCCACAUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAUAUGGGGCGGUGGC-CAGGAGUAU .....(((((((((.((.((((.((......))...)))).)).))---)...(((.(((.((((((........(((....)))......)))))).))).)))))-))))..... ( -34.64) >DroEre_CAF1 27761 108 + 1 ACCCACUUG----GAGUGGGAGAUAAGCGAGCAGGGCC-CGGCAUG---CGGGCACUGCAGCAUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAAGUGGGGCGGUGGC-CAGGAGUAU ..(((((((----..((((.(.....((..((((.(((-((.....---))))).)))).)).(((((........)))))...).)))).)))))))(((....))-)........ ( -40.60) >DroYak_CAF1 36980 112 + 1 ACCCACUUG----GAGUGGGAGAUAAACGAACUGGGCCUCGACAUUCGUCGGCCACUGCCACAUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAAGUGGGGCGGUGGC-CAGGAGUAU .(((((...----..))))).................((((((....)))((((((((((...(((((........))))).....((((....)))))))))))))-)..)))... ( -40.70) >DroAna_CAF1 26012 103 + 1 ACCCACUUGG----AGGGGGA----AGCG-GCAGACCUCGAACAUC---UCAACACU--CGCAUAUGAAAAUUAAAUUAUAAAAUGCUUUACAAGUGGGGCGGUGGCCCAGGAAUAU .(((((((((----((.((((----..((-(......)))....))---))....))--)((((((((........)))))...)))....))))))((((....)))).))..... ( -26.60) >consensus ACCCACUUGG___GAGUGGGAGAUGAACGAACAGGGCUCCGACACU___CGAGCACUGCCACAUAUGAAAAUUAAAUUAUAAAAUGCCAUGCAAGUGGGGCGGUGGC_CAGGAGUAU .(((((((.....)))))))...............((((((........)...(((((((...(((((........))))).....((((....))))))))))).....))))).. (-20.86 = -22.53 + 1.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:15 2006