
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
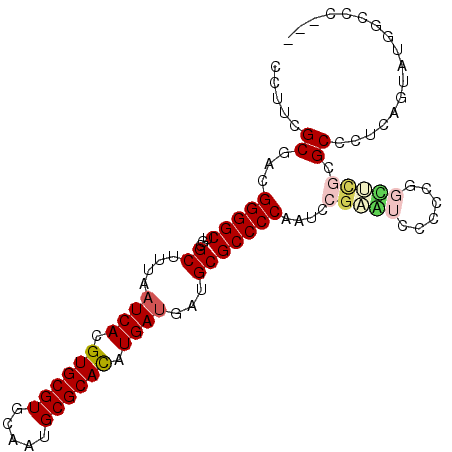
| Location | 21,643,746 – 21,643,844 |
| Length | 98 |
| Max. P | 0.774178 |

| Location | 21,643,746 – 21,643,844 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -27.28 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
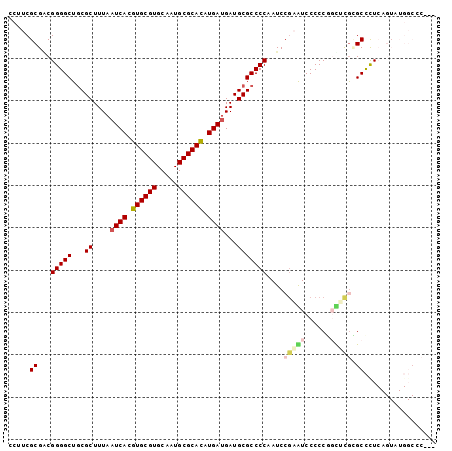
>3L_DroMel_CAF1 21643746 98 + 23771897 CCUUCGCGACGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUC---GUCCCCCGUUUCGCGCCCUCAGUAUGGCCCGAU .....((((..(((..((((....((((.((((((.....)))))).))))...)))))))..))---))........(((.(((........))).))). ( -36.50) >DroSec_CAF1 20422 101 + 1 CCAUCGCGCCGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAGUCCCCCGGCUCGCGCCCUCAGUAUGGCCCGAU ..((((.((((((((.((((....((((.((((((.....)))))).))))...)))).......((((((....)))))).)))))......))).)))) ( -43.50) >DroSim_CAF1 20689 101 + 1 CCAUCGCGACGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAGUCCCCCGGCUCGCGCCCUCAGUAUGGCCCGAU ((((.((...(((((.((((....((((.((((((.....)))))).))))...)))).......((((((....)))))).)))))..))))))...... ( -41.20) >DroEre_CAF1 19105 98 + 1 UCUUCGCGGCGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCAUAUGAUGAUGCGCCCCAAUCCGAAUCCCCCUGCUUGAGCCUUCAGUAUGGCUC--- .....((((.((((..((((....((((..(((((.....)))))..))))...))))............))))))))..(((((........)))))--- ( -38.14) >DroYak_CAF1 28581 87 + 1 CGUUCGCGGCGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCACAUGACGAUGCGCCCCAAUCCGAAUCCCCCUGUGCGAGCCCU-------------- .(((((((.(((((..((((.....(((.((((((.....)))))).)))....))))..(.....).....))))))))))))...-------------- ( -34.20) >DroPer_CAF1 8025 86 + 1 GUCUCGCGACGGGGCUACGCUUUAAUCACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGUAUCCCCCA-UGCAUGCUCU-------------- .....(((..(((((...((....((((.((((((.....)))))).))))...))))))).....((((.....)-))).)))...-------------- ( -29.50) >consensus CCUUCGCGACGGGGCUGCGCUUUAAUCACGUGCGUGCAAUGCGCACAUGAUGAUGCGCCCCAAUCCGAAUCCCCCGGCUCGCGCCCUCAGUAUGGCCC___ .....((...(((((...((....((((.((((((.....)))))).))))...)))))))....(((((......))))).))................. (-27.28 = -28.03 + 0.75)

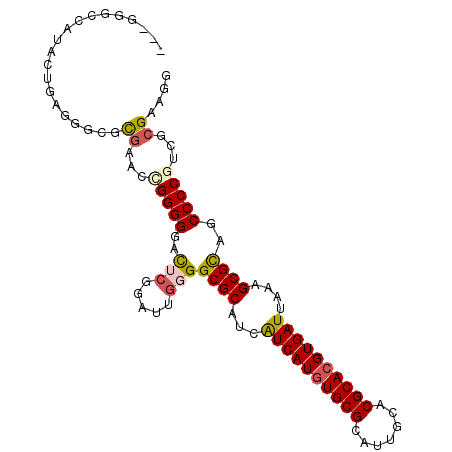
| Location | 21,643,746 – 21,643,844 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -27.60 |
| Energy contribution | -28.02 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
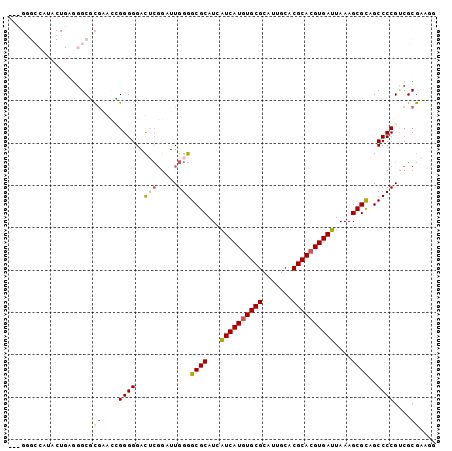
>3L_DroMel_CAF1 21643746 98 - 23771897 AUCGGGCCAUACUGAGGGCGCGAAACGGGGGAC---GAUUGGGGCGCAUCAUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGUCGCGAAGG .((((......))))...(((((..(((((..(---......)((((...((((((((((.......))))))))))....))))..)))))))))).... ( -40.50) >DroSec_CAF1 20422 101 - 1 AUCGGGCCAUACUGAGGGCGCGAGCCGGGGGACUCGGAUUGGGGCGCAUCAUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGGCGCGAUGG ((((.(((........(((....)))((((..(((.....)))((((...((((((((((.......))))))))))....))))..))))))).)))).. ( -50.30) >DroSim_CAF1 20689 101 - 1 AUCGGGCCAUACUGAGGGCGCGAGCCGGGGGACUCGGAUUGGGGCGCAUCAUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGUCGCGAUGG (((((((.........(((....)))((((..(((.....)))((((...((((((((((.......))))))))))....))))..))))))).)))).. ( -44.50) >DroEre_CAF1 19105 98 - 1 ---GAGCCAUACUGAAGGCUCAAGCAGGGGGAUUCGGAUUGGGGCGCAUCAUCAUAUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGCCGCGAAGA ---(((((........)))))..((.(((((..((......))((((...((((..((((.......))))..))))....))))..)))).).))..... ( -34.10) >DroYak_CAF1 28581 87 - 1 --------------AGGGCUCGCACAGGGGGAUUCGGAUUGGGGCGCAUCGUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGCCGCGAACG --------------...(.((((...((((...((......))((((...((((((((((.......))))))))))....))))..))))...)))).). ( -34.40) >DroPer_CAF1 8025 86 - 1 --------------AGAGCAUGCA-UGGGGGAUACGGAUUGGGGCGCAUCAUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGUAGCCCCGUCGCGAGAC --------------......((((-(((((..(((((((........)))((((((((((.......)))))))))).....)))).)))))).))).... ( -30.90) >consensus ___GGGCCAUACUGAGGGCGCGAACCGGGGGACUCGGAUUGGGGCGCAUCAUCAUGUGCGCAUUGCACGCACGUGAUUAAAGCGCAGCCCCGUCGCGAAGG ....................((...(((((..(((.....)))((((...((((((((((.......))))))))))....))))..)))))...)).... (-27.60 = -28.02 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:11 2006