
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
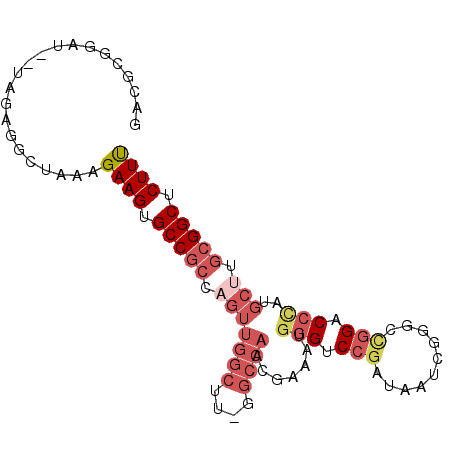
| Location | 21,643,191 – 21,643,283 |
| Length | 92 |
| Max. P | 0.940672 |

| Location | 21,643,191 – 21,643,283 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -19.65 |
| Energy contribution | -21.57 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21643191 92 + 23771897 GACGCGGAU--UGGAGGCUAAAGAAGUGCCGCCAGUUGGCUU-GGCCAACGCAAUGGGUCCGAUAAUCGGGCCGGACCCAUGCUUGCGGCUCUUU ...((.(((--(((.(((.........))).)))))).))..-((((.(.(((.((((((((..........))))))))))).)..)))).... ( -39.90) >DroSec_CAF1 19891 84 + 1 GACGCGGAU--UAGAGGCUAAAGAAGUGCCGCCA---------GGCCAACGAAAUGGGACCGAUAAUCGGGCCGGACCCAUGCCUGCGGCUCUUU ...((....--.....))....((((.(((((.(---------(((.......(((((.(((..........))).)))))))))))))).)))) ( -28.71) >DroSim_CAF1 20150 92 + 1 GACGCGGAU--UAGAGGCUAAAGAAGUGCCGCCAGUUGGCUU-GGCCAACGAAAUGGGACCGAUAAUCGGGCCGGACCCAUGCUUGCGGCUCUUU ...((....--.....))....((((.(((((..((((((..-.))))))...(((((.(((..........))).)))))....))))).)))) ( -31.50) >DroEre_CAF1 18565 89 + 1 -----AGAUUCUAGAGGAUGAGGAAGUGCCGCCAGUUGGCUU-GGCCAACGAAAUGGGUCCGAUAAUCGGGCCGGACCUCUGCUUGCGGCUCUUU -----..((((....))))...((((.(((((..((((((..-.))))))......((((((.....))))))............))))).)))) ( -33.10) >DroYak_CAF1 28043 83 + 1 -----------UAGAGGAUAAGGAAGUGCCGCCAGUUGGCUU-GGCCAAAAAAAUGGGUCCAAUAAUCUGGCCGGUCCUUUGCUUGCGGCUCUUU -----------...........((((.(((((.(((.((.((-(((((......((....))......))))))).))...))).))))).)))) ( -30.00) >DroAna_CAF1 17410 82 + 1 GAUGCGGAU--UAGAACCGAUAGAAGUGCCGACUGUUGGCCAAGGCCA--GAAAUGUGUCAG---------UUGGACCGUUGCUCGGGGCUCUUC ......((.--.((..(((((((..((.((((((((((((....))))--)........)))---------))))))..))).))))..))..)) ( -26.30) >consensus GACGCGGAU__UAGAGGCUAAAGAAGUGCCGCCAGUUGGCUU_GGCCAACGAAAUGGGUCCGAUAAUCGGGCCGGACCCAUGCUUGCGGCUCUUU ......................((((.(((((.(((((((....)))).......(((.(((..........))).)))..))).))))).)))) (-19.65 = -21.57 + 1.92)

| Location | 21,643,191 – 21,643,283 |
|---|---|
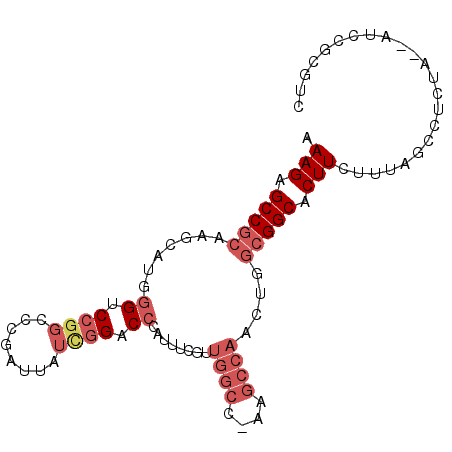
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -15.86 |
| Energy contribution | -17.42 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21643191 92 - 23771897 AAAGAGCCGCAAGCAUGGGUCCGGCCCGAUUAUCGGACCCAUUGCGUUGGCC-AAGCCAACUGGCGGCACUUCUUUAGCCUCCA--AUCCGCGUC ((((((((((..((((((((((((........))))))))).)))((((((.-..))))))..)))))...)))))........--......... ( -39.80) >DroSec_CAF1 19891 84 - 1 AAAGAGCCGCAGGCAUGGGUCCGGCCCGAUUAUCGGUCCCAUUUCGUUGGCC---------UGGCGGCACUUCUUUAGCCUCUA--AUCCGCGUC (((((((((((((((((((.((((........)))).))))).......)))---------).)))))...)))))........--......... ( -29.51) >DroSim_CAF1 20150 92 - 1 AAAGAGCCGCAAGCAUGGGUCCGGCCCGAUUAUCGGUCCCAUUUCGUUGGCC-AAGCCAACUGGCGGCACUUCUUUAGCCUCUA--AUCCGCGUC ((((((((((....(((((.((((........)))).)))))...((((((.-..))))))..)))))...)))))........--......... ( -32.20) >DroEre_CAF1 18565 89 - 1 AAAGAGCCGCAAGCAGAGGUCCGGCCCGAUUAUCGGACCCAUUUCGUUGGCC-AAGCCAACUGGCGGCACUUCCUCAUCCUCUAGAAUCU----- .(((.(((((.....(.(((((((........)))))))).....((((((.-..))))))..))))).)))..................----- ( -30.80) >DroYak_CAF1 28043 83 - 1 AAAGAGCCGCAAGCAAAGGACCGGCCAGAUUAUUGGACCCAUUUUUUUGGCC-AAGCCAACUGGCGGCACUUCCUUAUCCUCUA----------- ..((((.(....)..((((((((((((((..(.((....)).)..)))))))-..(((....)))))....)))))...)))).----------- ( -27.50) >DroAna_CAF1 17410 82 - 1 GAAGAGCCCCGAGCAACGGUCCAA---------CUGACACAUUUC--UGGCCUUGGCCAACAGUCGGCACUUCUAUCGGUUCUA--AUCCGCAUC ((((.((((((.....))).....---------..(((.......--((((....))))...)))))).))))...(((.....--..))).... ( -20.50) >consensus AAAGAGCCGCAAGCAUGGGUCCGGCCCGAUUAUCGGACCCAUUUCGUUGGCC_AAGCCAACUGGCGGCACUUCUUUAGCCUCUA__AUCCGCGUC .(((.(((((.......((.((((........)))).))........((((....))))....))))).)))....................... (-15.86 = -17.42 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:10:09 2006