
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,628,577 – 21,628,827 |
| Length | 250 |
| Max. P | 0.996786 |

| Location | 21,628,577 – 21,628,684 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21628577 107 + 23771897 AACAUAUAUGUACAGUAAUUAUUGGUU-UUUGACAAUAAAUGUAAAUUCGAUUAAGUUUCGAAAGGGUAGAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAU .....................(((((.-..((((............(((((.......)))))..((((....))))(....)............))))))))).... ( -19.60) >DroSec_CAF1 5035 107 + 1 AACAUAUAUGUACAGUAAUUAUUGGUU-UUUGACAAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAU .....................(((((.-..((((............(((((.......)))))..((((....))))(....)............))))))))).... ( -17.00) >DroSim_CAF1 5073 107 + 1 AACAUAUAUGUACAGUAAUUAUUGGUU-UUUGACAAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAU .....................(((((.-..((((............(((((.......)))))..((((....))))(....)............))))))))).... ( -17.00) >DroEre_CAF1 4185 107 + 1 AACAUAUAUGUACAGUAAUUAUUGGUU-UUUGACAAUAAAUGUAAAUUCAAUUAUGUUUCGAAAAGGUAAAAGUACCGGAAACAAAACCAAAAGAGUCAACCAAUAAU .................(((((((((.-..((((.((((.((......)).))))(((((.....((((....)))).)))))............))))))))))))) ( -19.50) >DroYak_CAF1 14964 108 + 1 AACAUAUAUGUACAGUAAUUAUUGGUUUUUUGACAAUAAAUGUAAAUUCAAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAGCCAAAAGAGUCAACCAAUAAU .................(((((((((....((((..(((.((......)).))).(((((.....((((....)))).)))))............))))))))))))) ( -18.30) >consensus AACAUAUAUGUACAGUAAUUAUUGGUU_UUUGACAAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAU .....................(((((....((((............(((((.......)))))..((((....))))(....)............))))))))).... (-14.80 = -15.36 + 0.56)

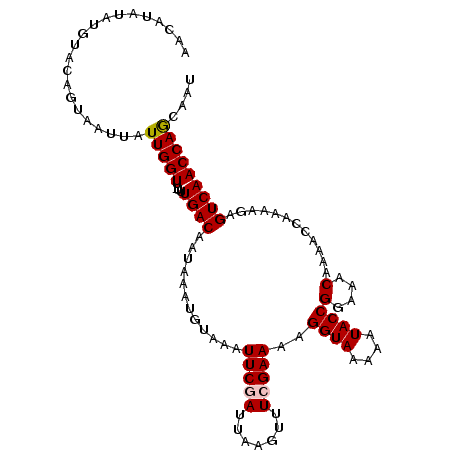
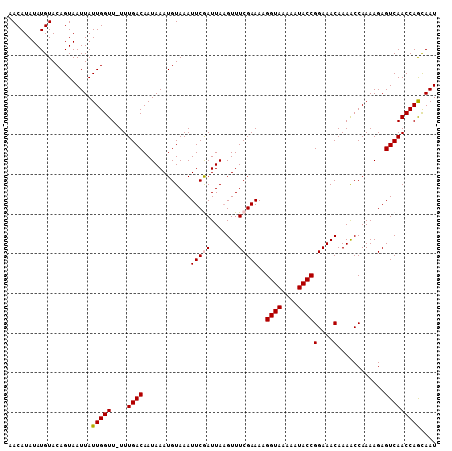
| Location | 21,628,577 – 21,628,684 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.06 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21628577 107 - 23771897 AUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUCUACCCUUUCGAAACUUAAUCGAAUUUACAUUUAUUGUCAAA-AACCAAUAAUUACUGUACAUAUAUGUU ((((.((((((((...(((((((..(((((.((((....)))).....)))))..)))))))............))))..-.)))).))))................. ( -16.76) >DroSec_CAF1 5035 107 - 1 AUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUUGUCAAA-AACCAAUAAUUACUGUACAUAUAUGUU ((((.((((((((...(((((((..(((((.((((....)))).....)))))..)))))))............))))..-.)))).))))................. ( -17.56) >DroSim_CAF1 5073 107 - 1 AUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUUGUCAAA-AACCAAUAAUUACUGUACAUAUAUGUU ((((.((((((((...(((((((..(((((.((((....)))).....)))))..)))))))............))))..-.)))).))))................. ( -17.56) >DroEre_CAF1 4185 107 - 1 AUUAUUGGUUGACUCUUUUGGUUUUGUUUCCGGUACUUUUACCUUUUCGAAACAUAAUUGAAUUUACAUUUAUUGUCAAA-AACCAAUAAUUACUGUACAUAUAUGUU (((((((((((((...((..(((.((((((.((((....)))).....)))))).)))..))............))))..-.)))))))))................. ( -20.26) >DroYak_CAF1 14964 108 - 1 AUUAUUGGUUGACUCUUUUGGCUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUUGAAUUUACAUUUAUUGUCAAAAAACCAAUAAUUACUGUACAUAUAUGUU ((((((((((.....(((((((...(((((.((((....)))).....))))).....(((((....)))))..)))))))))))))))))................. ( -18.80) >consensus AUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUUGUCAAA_AACCAAUAAUUACUGUACAUAUAUGUU ((((.((((((((...(((((((..(((((.((((....)))).....)))))..)))))))............))))....)))).))))................. (-15.34 = -15.06 + -0.28)


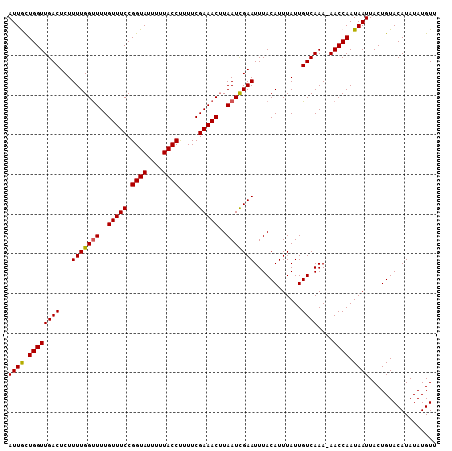
| Location | 21,628,610 – 21,628,722 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.16 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21628610 112 + 23771897 AAUAAAUGUAAAUUCGAUUAAGUUUCGAAAGGGUAGAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAUGAAAGAGGUUCCGAAUGAUUGAUGAACAUGAAGGUAAU .....((((....(((((((...((((....((((....))))(....)..((((.......(((........)))....)))).)))))))))))...))))......... ( -18.50) >DroSec_CAF1 5068 112 + 1 AAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAUGAAAAAGGUUCCGAAUGAUUGAUGAACAUGAAGGUAAU .....((((....(((((((...((((....((((....))))(....)..((((.......(((........)))....)))).)))))))))))...))))......... ( -15.90) >DroSim_CAF1 5106 112 + 1 AAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAUGAAAAAGGUUCCGAAUGAUUGAUGAACAUGAAGGUAAU .....((((....(((((((...((((....((((....))))(....)..((((.......(((........)))....)))).)))))))))))...))))......... ( -15.90) >DroEre_CAF1 4218 112 + 1 AAUAAAUGUAAAUUCAAUUAUGUUUCGAAAAGGUAAAAGUACCGGAAACAAAACCAAAAGAGUCAACCAAUAAUGAAAGAGGUGCCGAGUGAUUGAUGAACAUAAAGGUAAU .....((((....(((((((((((((.....((((....)))).)))))...(((.......(((........)))....))).....))))))))...))))......... ( -16.80) >DroYak_CAF1 14998 112 + 1 AAUAAAUGUAAAUUCAAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAGCCAAAAGAGUCAACCAAUAAUGAAAUAGGUUCCGAGUGAUUGAUGAACAUAAGGGUAAU .....((((....(((((((...((((....((((....))))(....)..((((.......(((........)))....)))).)))))))))))...))))......... ( -15.90) >consensus AAUAAAUGUAAAUUCGAUUAAGUUUCGAAAAGGUAAAAAUACCGGAAACAAAACCAAAAGAGUCAACCAGCAAUGAAAAAGGUUCCGAAUGAUUGAUGAACAUGAAGGUAAU .....((((....(((((((...((((....((((....))))(....)..((((.......(((........)))....)))).)))))))))))...))))......... (-16.20 = -16.16 + -0.04)

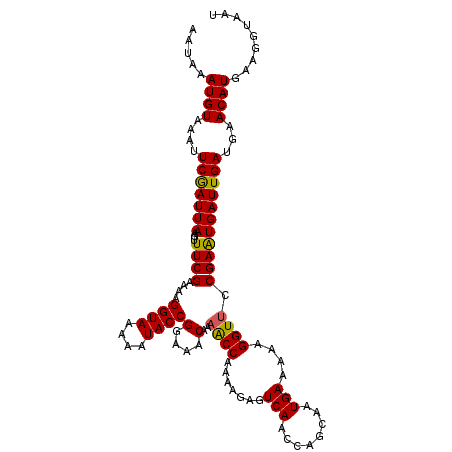

| Location | 21,628,610 – 21,628,722 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -16.89 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21628610 112 - 23771897 AUUACCUUCAUGUUCAUCAAUCAUUCGGAACCUCUUUCAUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUCUACCCUUUCGAAACUUAAUCGAAUUUACAUUUAUU .........((((...(((((((....(((.....))).....)))))))....(((((((..(((((.((((....)))).....)))))..)))))))...))))..... ( -17.90) >DroSec_CAF1 5068 112 - 1 AUUACCUUCAUGUUCAUCAAUCAUUCGGAACCUUUUUCAUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUU .........((((...(((((((....(((.....))).....)))))))....(((((((..(((((.((((....)))).....)))))..)))))))...))))..... ( -18.70) >DroSim_CAF1 5106 112 - 1 AUUACCUUCAUGUUCAUCAAUCAUUCGGAACCUUUUUCAUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUU .........((((...(((((((....(((.....))).....)))))))....(((((((..(((((.((((....)))).....)))))..)))))))...))))..... ( -18.70) >DroEre_CAF1 4218 112 - 1 AUUACCUUUAUGUUCAUCAAUCACUCGGCACCUCUUUCAUUAUUGGUUGACUCUUUUGGUUUUGUUUCCGGUACUUUUACCUUUUCGAAACAUAAUUGAAUUUACAUUUAUU .........((((...(((((((....................)))))))....((..(((.((((((.((((....)))).....)))))).)))..))...))))..... ( -14.85) >DroYak_CAF1 14998 112 - 1 AUUACCCUUAUGUUCAUCAAUCACUCGGAACCUAUUUCAUUAUUGGUUGACUCUUUUGGCUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUUGAAUUUACAUUUAUU ...........((((((((((((....(((.....))).....))))))).............(((((.((((....)))).....))))).....)))))........... ( -14.30) >consensus AUUACCUUCAUGUUCAUCAAUCAUUCGGAACCUCUUUCAUUGCUGGUUGACUCUUUUGGUUUUGUUUCCGGUAUUUUUACCUUUUCGAAACUUAAUCGAAUUUACAUUUAUU .........((((...(((((((....(((.....))).....)))))))....(((((((..(((((.((((....)))).....)))))..)))))))...))))..... (-15.62 = -15.78 + 0.16)



| Location | 21,628,722 – 21,628,827 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -22.26 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21628722 105 + 23771897 GAAAAAAUUACAGAUACCACACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGGGUCGUGAGCGUGUCGAGAAUUUUUUGAUUACGUUUGACAAGGUUUGAA ..........(((((.((.((((..((.((.(((......))))).))..)))).))(((..(((((((((((((...)))))).))))))))))...))))).. ( -24.70) >DroSec_CAF1 5180 105 + 1 GAAAAAAUUACAGAUACCACACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGGGUCGUGAGCGUGUCGAGAAUUUUUUGAUUACGUUUGACAAGGUUUGAA ..........(((((.((.((((..((.((.(((......))))).))..)))).))(((..(((((((((((((...)))))).))))))))))...))))).. ( -24.70) >DroSim_CAF1 5218 105 + 1 GAAAAAAUUACAGAUACCACACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGGGUCGUGAGCGUGUCGAGAAUUUUUUGAUUACGUUUGACAAGGUUUGAA ..........(((((.((.((((..((.((.(((......))))).))..)))).))(((..(((((((((((((...)))))).))))))))))...))))).. ( -24.70) >DroEre_CAF1 4330 105 + 1 GAAAAAAUUACAGAAACCACACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGAGUCGUGAGCGUGUCGAGAAUUUUUUGAUUAUGUUUGACAAGGUUUGAG .............(((((.((((..((.((.(((......))))).))..))))...(((..(((((((((((((...)))))).))))))))))..)))))... ( -20.70) >DroYak_CAF1 15110 105 + 1 GAAAAAAUUACAGAUACCGCACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGAGUCGUGAGCGUGUCGAGAAUUUUUUGAUUACGUUCGACAAGGUUUGAG ..........(((((..((((((..((.((.(((......))))).))..)))))).(((..(((((((((((((...)))))).))))))))))...))))).. ( -28.20) >consensus GAAAAAAUUACAGAUACCACACGAAUGGAUUAUGAAAUGACAUAUGCAAGUGUGCGGGUCGUGAGCGUGUCGAGAAUUUUUUGAUUACGUUUGACAAGGUUUGAA ..........(((((.((((((...((.((.(((......))))).)).))))).).(((..(((((((((((((...)))))).))))))))))...))))).. (-22.26 = -21.98 + -0.28)

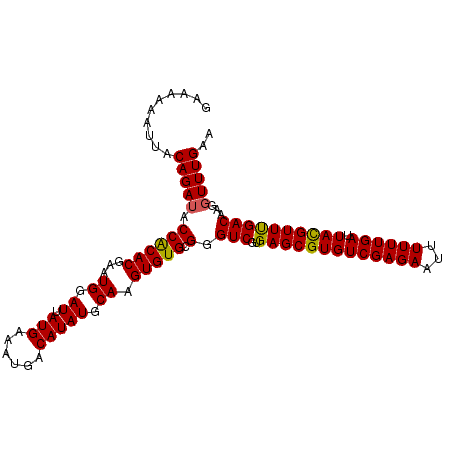
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:56 2006