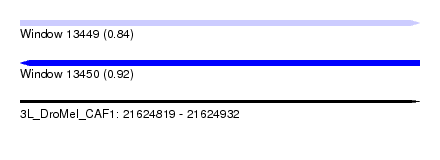
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,624,819 – 21,624,932 |
| Length | 113 |
| Max. P | 0.922145 |

| Location | 21,624,819 – 21,624,932 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21624819 113 + 23771897 GACGAGAAAGGGCGCUGGUUGAGCCAUGAUGAAAUUGUUGGCUUCGAUUUUGGCUAAUUGCCAGAGUCCAUUUGA-------AAUGCCCACUUAGGGCUAAUUGGUCCCCUCGGAAGCGC ..((((...(((.((..(((((((((............)))))).(((((((((.....))))))))).......-------...((((.....)))).)))..)))))))))....... ( -39.60) >DroSec_CAF1 1173 113 + 1 GACGAGGAAGGGCGCUGGUUGAGCCAUGAUGAAAUUGUUGGCUUCGAUUUUGGCUAAUUGCCGGAGUCCAUUUGA-------AAUGCCCACUUAGGGCUAAUUGGUCCCCUCGGAAGCGC ..(((((..(((((......((((((............)))))).(((((((((.....))))))))).......-------..))))).....(((((....))))))))))....... ( -41.30) >DroSim_CAF1 1161 113 + 1 GACGAGGAAGGGCGCUGGUUGAGCCAUGAUGAAAUUGUUGGCUUCAAUUUUGGCUAAUUGCCGGAGUCCAUUUGA-------AAUGCCCACUUAGAGCUAAUUGGUCCCCUCGGAAGCGC ..(((((..((((.(((((..(((((.....((((((.......)))))))))))....))))).))))......-------.....(((.(((....))).)))...)))))....... ( -34.60) >DroEre_CAF1 143 113 + 1 GACGAGGAAGGGCGAUGGCUGAGCCAUGAUGAAAUUGUUGGCUUCGAUUUUGGCUAAUUGCCGGAGUCCAUUUGA-------AAUGCCCACUUAGGGCUAAUUGGUCCCCUCGGAAGCGC ..(((((..(((((......((((((............)))))).(((((((((.....))))))))).......-------..))))).....(((((....))))))))))....... ( -41.60) >DroYak_CAF1 10586 120 + 1 GAUGUGUAAGGGCGAUGGUUGAGCCAUGAUGAAAUUGUUGGCUUCGAUUUUGGCUAAUUGCCGGAGUCCAUUUGAAAUGUGAAAUGCCCACUUAGGGCUAAUUGGUCCCCUCGGAAGCGC ...((((..(((((......((((((............)))))).(((((((((.....)))))))))................))))).....(((((....)))))........)))) ( -35.10) >consensus GACGAGGAAGGGCGCUGGUUGAGCCAUGAUGAAAUUGUUGGCUUCGAUUUUGGCUAAUUGCCGGAGUCCAUUUGA_______AAUGCCCACUUAGGGCUAAUUGGUCCCCUCGGAAGCGC ..(((((..(((((......((((((............)))))).(((((((((.....)))))))))................))))).....(((((....))))))))))....... (-35.72 = -36.40 + 0.68)



| Location | 21,624,819 – 21,624,932 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21624819 113 - 23771897 GCGCUUCCGAGGGGACCAAUUAGCCCUAAGUGGGCAUU-------UCAAAUGGACUCUGGCAAUUAGCCAAAAUCGAAGCCAACAAUUUCAUCAUGGCUCAACCAGCGCCCUUUCUCGUC (((((..(((((((.(((....((((.....))))...-------.....))).))))(((.....)))....))).(((((............))))).....)))))........... ( -30.10) >DroSec_CAF1 1173 113 - 1 GCGCUUCCGAGGGGACCAAUUAGCCCUAAGUGGGCAUU-------UCAAAUGGACUCCGGCAAUUAGCCAAAAUCGAAGCCAACAAUUUCAUCAUGGCUCAACCAGCGCCCUUCCUCGUC (((((..(((((((.(((....((((.....))))...-------.....))).))))(((.....)))....))).(((((............))))).....)))))........... ( -33.00) >DroSim_CAF1 1161 113 - 1 GCGCUUCCGAGGGGACCAAUUAGCUCUAAGUGGGCAUU-------UCAAAUGGACUCCGGCAAUUAGCCAAAAUUGAAGCCAACAAUUUCAUCAUGGCUCAACCAGCGCCCUUCCUCGUC (((((.....((((.(((....((((.....))))...-------.....))).))))(((.....)))....((((.((((............))))))))..)))))........... ( -29.40) >DroEre_CAF1 143 113 - 1 GCGCUUCCGAGGGGACCAAUUAGCCCUAAGUGGGCAUU-------UCAAAUGGACUCCGGCAAUUAGCCAAAAUCGAAGCCAACAAUUUCAUCAUGGCUCAGCCAUCGCCCUUCCUCGUC (((((((.((((((.(((....((((.....))))...-------.....))).))))(((.....)))....))))))).............(((((...))))).))........... ( -31.60) >DroYak_CAF1 10586 120 - 1 GCGCUUCCGAGGGGACCAAUUAGCCCUAAGUGGGCAUUUCACAUUUCAAAUGGACUCCGGCAAUUAGCCAAAAUCGAAGCCAACAAUUUCAUCAUGGCUCAACCAUCGCCCUUACACAUC (((((((.((((((.((.....((((.....))))((((........)))))).))))(((.....)))....))))))).............((((.....)))).))........... ( -29.60) >consensus GCGCUUCCGAGGGGACCAAUUAGCCCUAAGUGGGCAUU_______UCAAAUGGACUCCGGCAAUUAGCCAAAAUCGAAGCCAACAAUUUCAUCAUGGCUCAACCAGCGCCCUUCCUCGUC ........((((((........((((.....))))...............(((.....(((.....)))......((.((((............))))))..)))...))))))...... (-28.46 = -28.38 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:47 2006