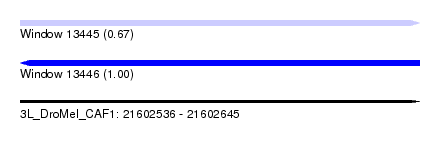
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,602,536 – 21,602,645 |
| Length | 109 |
| Max. P | 0.998265 |

| Location | 21,602,536 – 21,602,645 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21602536 109 + 23771897 ACCUGCAAAUCGCUGAGCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUGGAAUUGAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUG-U ....(((((((..((((....))))((((........(((((((((((..(((.((((..........)))).))).))).))))))))........))))).)))))-) ( -24.59) >DroVir_CAF1 38324 103 + 1 ------GAUUGGUUGAACAAAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGCUC-AAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAACUUUAU ------..((((((((...........((....))(.(((((((((((..(((.((((....-.....)))).))).))).)))))))).)...))))))))........ ( -23.90) >DroPse_CAF1 39276 106 + 1 GGUUG---UUUGUUGGAUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGAUCGAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUU-G (((((---((((((((......)))............(((((((((((..(((.((((..........)))).))).))).))))))))))))).)))))........-. ( -23.10) >DroGri_CAF1 33393 103 + 1 ------GGUUUGUUGAUUACAUUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGCUC-AAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAACUCUGA ------((((((((((((.........((....))(.(((((((((((..(((.((((....-.....)))).))).))).)))))))).).)))))))..))))).... ( -24.30) >DroWil_CAF1 48142 106 + 1 GAG---AAUUUGUUGAUUAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUAGUUAAAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUA-U ..(---((((((((((((.........((....))(.(((((((((((..(((.((((..........)))).))).))).)))))))).).)))))))..)))))).-. ( -23.20) >DroAna_CAF1 34065 104 + 1 -----CGAAAUGUUGACCAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGUUUGAAUUAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUG-A -----............((((.((.((((........(((((((((((..(((.((((..........)))).))).))).))))))))........)))))).))))-. ( -22.89) >consensus ______GAUUUGUUGAACAAACUCAUAGUAAAUACGCCUUUUUCCCUUUCCUGUCGUUGCUC_AAUAAAACGCCAGUAAGCGGAAAAAGACAAAUCGACUAGAAUUUU_U ...........(((((...........((....))(.(((((((((((..(((.((((..........)))).))).))).)))))))).)...)))))........... (-19.72 = -19.75 + 0.03)


| Location | 21,602,536 – 21,602,645 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -30.26 |
| Energy contribution | -29.85 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21602536 109 - 23771897 A-CAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUCAAUUCCACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGCUCAGCGAUUUGCAGGU .-(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))....((.....)).... ( -33.30) >DroVir_CAF1 38324 103 - 1 AUAAAGUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUU-GAGCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUUUGUUCAACCAAUC------ ...((((((((((.(((.(((((((((((.(((.(((.((((.....-....)))).)))..)))))))))))))).))).)))).))))))............------ ( -30.10) >DroPse_CAF1 39276 106 - 1 C-AAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUCGAUCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAUCCAACAAA---CAACC .-.((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).))))))..........---..... ( -30.00) >DroGri_CAF1 33393 103 - 1 UCAGAGUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUU-GAGCAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAAUGUAAUCAACAAACC------ .....((((((((.(((.(((((((((((.(((.(((.((((.....-....)))).)))..)))))))))))))).))).)))).))))..............------ ( -28.60) >DroWil_CAF1 48142 106 - 1 A-UAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUUUAACUAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAAUCAACAAAUU---CUC .-(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))))...........---... ( -32.10) >DroAna_CAF1 34065 104 - 1 U-CAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUAAUUCAAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUGGUCAACAUUUCG----- (-(((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).))))))))...........----- ( -33.20) >consensus A_AAAAUUCUAGUCGAUUUGUCUUUUUCCGCUUACUGGCGUUUUAUU_GAACAACGACAGGAAAGGGAAAAAGGCGUAUUUACUAUGAGUUUAAUCAACAAAUC______ ...((((((((((.(((.(((((((((((.(((.(((.((((..........)))).)))..)))))))))))))).))).)))).)))))).................. (-30.26 = -29.85 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:43 2006