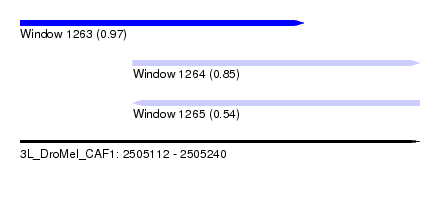
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,505,112 – 2,505,240 |
| Length | 128 |
| Max. P | 0.974062 |

| Location | 2,505,112 – 2,505,203 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -16.12 |
| Energy contribution | -17.36 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2505112 91 + 23771897 GAUUGCGCUCGAAUUACACUUAACACAAAUUUCAUUUGAAUAAACUAAAUG---------GCCAAAGGAGACGGGCUGCAUAAUCAGCAAAAAAAAAAAA (((((.((........................((((((.......))))))---------(((...(....).))).)).)))))............... ( -15.20) >DroSec_CAF1 128211 91 + 1 GAUUGCGCUCGAAUUACACUUAACACAAUUUUCAUUUGAAUAAACUAAAUGAAAUGCACGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA--------- ..((((........................((((((((.......))))))))(((((..(((...(....).)))))))).....)))).--------- ( -20.60) >DroSim_CAF1 123546 90 + 1 GAUUGCGCUCGAA-UACACUUAACACAAAUUUCAUUUGAAUAAACUAAAUGGAAUGCACGGCCAAAGGAGACGGGAUGCAUAAUCAGCAAA--------- ..((((.......-................((((((((.......))))))))(((((...((...(....).)).))))).....)))).--------- ( -17.70) >DroEre_CAF1 128763 91 + 1 GAUUGCGCUCGAAUUACACUUAACACAAAUUUCAUUUGAAUAAACUAAAUGAAAUGCAUGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA--------- (((((.((....................((((((((((.......))))))))))....((((...(....).)))))).)))))......--------- ( -20.90) >DroYak_CAF1 127198 91 + 1 GAUUGCGCUCGAAUUACACUUAACACAAAUUUCAUUUGAAUAAACUAAAUGGAAUGCAUGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA--------- (((((.((....................((((((((((.......))))))))))....((((...(....).)))))).)))))......--------- ( -20.40) >consensus GAUUGCGCUCGAAUUACACUUAACACAAAUUUCAUUUGAAUAAACUAAAUGAAAUGCACGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA_________ (((((.((....................((((((((((.......))))))))))....((((...(....).)))))).)))))............... (-16.12 = -17.36 + 1.24)

| Location | 2,505,148 – 2,505,240 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -16.84 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2505148 92 + 23771897 UGAAUAAACUAAAUG---------GCCAAAGGAGACGGGCUGCAUAAUCAGCAAAAAAAAAAAAAAAACUCAUUAAAAUGUUUAAUCAGGAAAAUGGGUGG ..............(---------(((...(....).))))((.......))...............(((((((....((......))....))))))).. ( -15.20) >DroSec_CAF1 128247 91 + 1 UGAAUAAACUAAAUGAAAUGCACGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA----------AAACUCAUUAAAAUGUUUAAUCAGGAAAAUGGGUGG .............(((.(((((..(((...(....).))))))))..))).....----------..(((((((....((......))....))))))).. ( -18.60) >DroSim_CAF1 123581 91 + 1 UGAAUAAACUAAAUGGAAUGCACGGCCAAAGGAGACGGGAUGCAUAAUCAGCAAA----------AAACUCAUUAAAAUGUUUAAUCAGGAAAAUGGGUGG .................(((((...((...(....).)).)))))..........----------..(((((((....((......))....))))))).. ( -15.50) >DroEre_CAF1 128799 91 + 1 UGAAUAAACUAAAUGAAAUGCAUGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA----------AAACUCAUUAAAAUGUUUAAUCACGAAAAUGGGUUG .............(((.(((((..(((...(....).))))))))..))).....----------.((((((((....((......))....)))))))). ( -18.40) >DroYak_CAF1 127234 91 + 1 UGAAUAAACUAAAUGGAAUGCAUGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA----------AAACUCAUUAAAAUGUUUAAUCACGAAAAUGGCUGG .................(((((..(((...(....).))))))))...((((...----------......(((((.....)))))..........)))). ( -16.51) >consensus UGAAUAAACUAAAUGAAAUGCACGGCCAAAGGAGACGGGCUGCAUAAUCAGCAAA__________AAACUCAUUAAAAUGUUUAAUCAGGAAAAUGGGUGG (((.(((((..(((((........(((...(....).)))(((.......)))................))))).....))))).)))............. (-13.34 = -13.58 + 0.24)

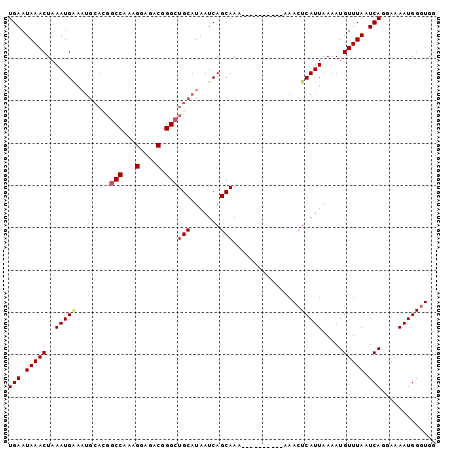
| Location | 2,505,148 – 2,505,240 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -14.70 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2505148 92 - 23771897 CCACCCAUUUUCCUGAUUAAACAUUUUAAUGAGUUUUUUUUUUUUUUUUGCUGAUUAUGCAGCCCGUCUCCUUUGGC---------CAUUUAGUUUAUUCA ..............((.(((((...........................((((......))))..(((......)))---------......))))).)). ( -11.50) >DroSec_CAF1 128247 91 - 1 CCACCCAUUUUCCUGAUUAAACAUUUUAAUGAGUUU----------UUUGCUGAUUAUGCAGCCCGUCUCCUUUGGCCGUGCAUUUCAUUUAGUUUAUUCA ..............((.(((((....((((.(((..----------...))).))))(((((((.(......).)))..)))).........))))).)). ( -15.30) >DroSim_CAF1 123581 91 - 1 CCACCCAUUUUCCUGAUUAAACAUUUUAAUGAGUUU----------UUUGCUGAUUAUGCAUCCCGUCUCCUUUGGCCGUGCAUUCCAUUUAGUUUAUUCA ..............((.(((((....((((.(((..----------...))).))))(((((...(((......))).))))).........))))).)). ( -15.90) >DroEre_CAF1 128799 91 - 1 CAACCCAUUUUCGUGAUUAAACAUUUUAAUGAGUUU----------UUUGCUGAUUAUGCAGCCCGUCUCCUUUGGCCAUGCAUUUCAUUUAGUUUAUUCA .............(((.(((((....((((.(((..----------...))).))))(((((((.(......).)))..)))).........))))).))) ( -15.40) >DroYak_CAF1 127234 91 - 1 CCAGCCAUUUUCGUGAUUAAACAUUUUAAUGAGUUU----------UUUGCUGAUUAUGCAGCCCGUCUCCUUUGGCCAUGCAUUCCAUUUAGUUUAUUCA .............(((.(((((....((((.(((..----------...))).))))(((((((.(......).)))..)))).........))))).))) ( -15.40) >consensus CCACCCAUUUUCCUGAUUAAACAUUUUAAUGAGUUU__________UUUGCUGAUUAUGCAGCCCGUCUCCUUUGGCCAUGCAUUCCAUUUAGUUUAUUCA .............(((.(((((....((((.(((...............))).))))((((..(((.......)))...)))).........))))).))) (-10.80 = -11.60 + 0.80)

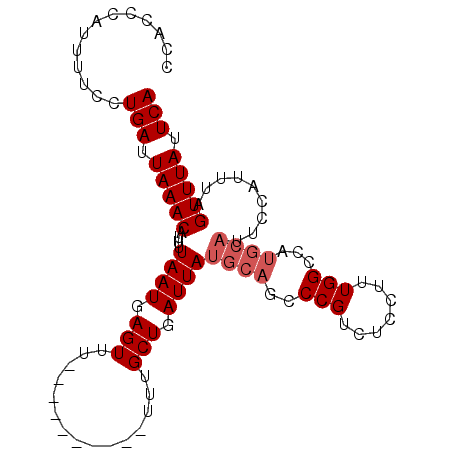
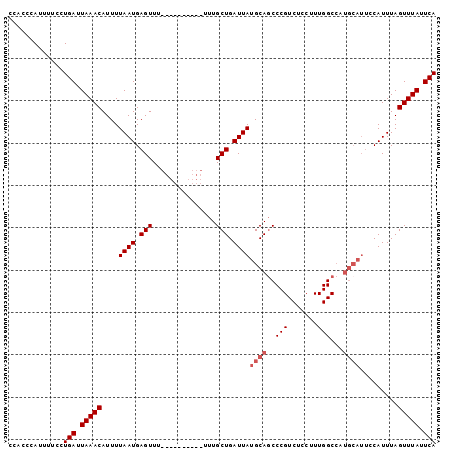
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:03 2006