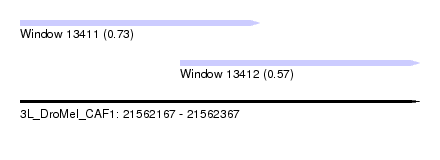
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,562,167 – 21,562,367 |
| Length | 200 |
| Max. P | 0.726816 |

| Location | 21,562,167 – 21,562,287 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
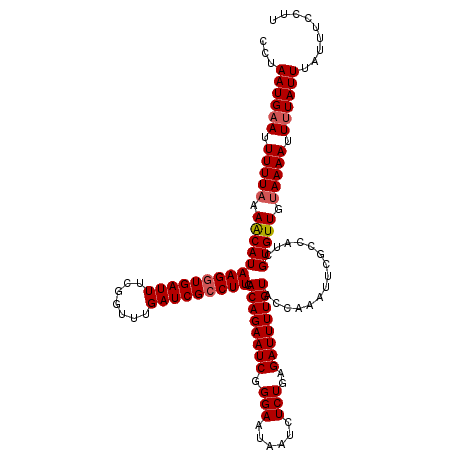
>3L_DroMel_CAF1 21562167 120 + 23771897 AACUGAAUACAUUUAUUAUACUCUACAACAUAUUUAGAGCGAUUUUAUCGUACAUAUCGCAUCGCUUUCCUUAAGAUUUACCUAAUGCAUUUUUAAAGCAUAAGGUGAUUUCGGUUUGAU (((((((............................((((((((.....((.......)).)))))))).........((((((.((((.........)))).)))))).))))))).... ( -21.80) >DroSec_CAF1 9828 120 + 1 AACUGAAUACAUUUAUUAUACUCUACAACAUAUUUAGAGCGAUGUUAUCGUACAUAUCGCAUCGCUUUCCUUAAGAUUUACCUAAUGAAUUUUUAAAACAUAAGGUGAUUUCAGUUUGAU (((((((............................((((((((((.((.......)).)))))))))).........((((((.(((...........))).)))))).))))))).... ( -23.30) >DroSim_CAF1 9997 120 + 1 AACUGAAUACAUUUAUUAUACUCUACAACAUAUUUAGAGCGAUGUUAUCGUACAUAUCGCAUCGCUUUCCUUAAGAUUUACCUAAUGAAUUUUUUAAACAUAAGGUGAUUUCGGUUUGAU (((((((............................((((((((((.((.......)).)))))))))).........((((((.(((...........))).)))))).))))))).... ( -22.60) >consensus AACUGAAUACAUUUAUUAUACUCUACAACAUAUUUAGAGCGAUGUUAUCGUACAUAUCGCAUCGCUUUCCUUAAGAUUUACCUAAUGAAUUUUUAAAACAUAAGGUGAUUUCGGUUUGAU (((((((............................((((((((((.((.......)).)))))))))).........((((((.(((...........))).)))))).))))))).... (-21.62 = -21.73 + 0.11)


| Location | 21,562,247 – 21,562,367 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -23.72 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21562247 120 + 23771897 CCUAAUGCAUUUUUAAAGCAUAAGGUGAUUUCGGUUUGAUCGACUUAACAGAAUCGGGAAUAAUCUCUGAGAUUUUGUACCAAAUUCGCCAUCGUGUUGUAAAAUUUUAUUUAUUUCCUU ..........(((((.(((((..(((((.((.(((............((((((((.(((......)))..))))))))))).)).)))))...))))).)))))................ ( -22.40) >DroSec_CAF1 9908 120 + 1 CCUAAUGAAUUUUUAAAACAUAAGGUGAUUUCAGUUUGAUCGCCUUCACAGAAUCGGGAAUAAUCUCUGAGAUUUUGUACCAAAUUCGCCAUCGUGUUGUAAAAUUUUAUUUAUUUCCUU ...((((((.(((((.((((((((((((((.......))))))))).((((((((.(((......)))..))))))))...............))))).))))).))))))......... ( -27.40) >DroSim_CAF1 10077 120 + 1 CCUAAUGAAUUUUUUAAACAUAAGGUGAUUUCGGUUUGAUCGCCUUCACAGAAUCGGGAAUAAUCUCUGAGAUUUUGUACCAAAUUCGCCAUCGUGUUGUAAAAUUUUAUUUAUUUCCUU ......(((((((...(((((..(((((.((.(((..((......))((((((((.(((......)))..))))))))))).)).)))))...)))))..)))))))............. ( -25.30) >consensus CCUAAUGAAUUUUUAAAACAUAAGGUGAUUUCGGUUUGAUCGCCUUCACAGAAUCGGGAAUAAUCUCUGAGAUUUUGUACCAAAUUCGCCAUCGUGUUGUAAAAUUUUAUUUAUUUCCUU ...((((((.(((((.((((((((((((((.......))))))))).((((((((.(((......)))..))))))))...............))))).))))).))))))......... (-23.72 = -24.50 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:10 2006