| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,545,488 – 21,545,596 |
| Length | 108 |
| Max. P | 0.961171 |

| Location | 21,545,488 – 21,545,596 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -14.45 |
| Energy contribution | -16.12 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
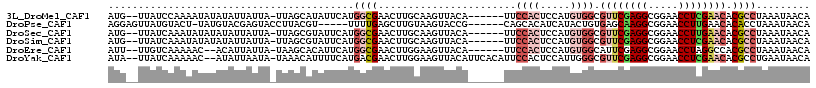
>3L_DroMel_CAF1 21545488 108 + 23771897 AUG--UUAUCCAAAAUAUAUAUUAUUA-UUAGCAUAUUCAUGGCGAACUUGCAAGUUACA------UUCCACUCCAUGUGGCGUUCGAGGCGGAACCUCGAACACGCCUAAAUAACA (((--(((.....((((.....)))).-.))))))......((((.........((((((------(........)))))))((((((((.....)))))))).))))......... ( -28.50) >DroPse_CAF1 46308 105 + 1 AGGAGUUAUGUACU-UAUGUACGAGUACCUUACGU-----UUUUGAGCUUGUAAGUACCG------CAGCACAUCAUACUGUGAGCAAGGCGGAACCUUGAACACACCUAAAUAACA (((.(((..(((((-(((((.((((.((.....))-----.)))).))..))))))))..------.))).........((((..(((((.....)))))..)))))))........ ( -23.50) >DroSec_CAF1 17204 108 + 1 AUG--UUAUCAAAUAUAUAUAUUAUUA-UUAGCGUAUUCAUGGCGAACUUGCAAGUUACA------UUCCACUCCAUGUGGCGUUCGAGGCGGAACCUUGAACACGCCUAAAUAACA (((--(((...((((.......)))).-.))))))......((((.........((((((------(........)))))))((((((((.....)))))))).))))......... ( -27.10) >DroSim_CAF1 20985 108 + 1 AUG--UUAUCAAAUAUAUAUAUUAUUA-UUAGCGUAUUCAUGGCGAACUUGCAAGUUACA------UUCCACUCCAUGUGGCGUUCGAGGCGGAACCUCGAACACGCCUAAAUAACA (((--(((...((((.......)))).-.))))))......((((.........((((((------(........)))))))((((((((.....)))))))).))))......... ( -29.20) >DroEre_CAF1 35156 106 + 1 AUU--UUGUCAAAAAC--ACAUUAUUA-UAAGCACAUUCAUGGCGAACUUGGAAGUUACA------UUCCACUCCAUGUGGCAUUCGAGGCGGAACCUAGGCCACGCCUAAAUAACA ...--...........--.........-.............(((.....(((((......------)))))......(((((.....(((.....)))..))))))))......... ( -21.30) >DroYak_CAF1 35471 112 + 1 AUA--UUAUCAAAAAC--AUAUUAAUA-UAAACAUUUUCAUGACGAACUUGGAAGUUACAUUCACAUUCCACUCCAUUGGGCGUUCGAGGCGGAACCUCGAACACGCCUGAAUAACA (((--(((........--....)))))-)....................(((((((.......)).))))).....(..(((((((((((.....))))))..)))))..)...... ( -23.60) >consensus AUG__UUAUCAAAAAUAUAUAUUAUUA_UUAGCAUAUUCAUGGCGAACUUGCAAGUUACA______UUCCACUCCAUGUGGCGUUCGAGGCGGAACCUCGAACACGCCUAAAUAACA .........................................((((.......................((((.....)))).((((((((.....)))))))).))))......... (-14.45 = -16.12 + 1.67)

| Location | 21,545,488 – 21,545,596 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -17.22 |
| Energy contribution | -18.51 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21545488 108 - 23771897 UGUUAUUUAGGCGUGUUCGAGGUUCCGCCUCGAACGCCACAUGGAGUGGAA------UGUAACUUGCAAGUUCGCCAUGAAUAUGCUAA-UAAUAAUAUAUAUUUUGGAUAA--CAU ((((((((((((((((((((((.....)))))))).((((.....)))).)------))).....(((.((((.....)))).)))...-..............))))))))--)). ( -32.80) >DroPse_CAF1 46308 105 - 1 UGUUAUUUAGGUGUGUUCAAGGUUCCGCCUUGCUCACAGUAUGAUGUGCUG------CGGUACUUACAAGCUCAAAA-----ACGUAAGGUACUCGUACAUA-AGUACAUAACUCCU .(((((.....((((..(((((.....)))))..))))((((..((((.((------((((((((((..........-----..)))).)))).))))))))-.))))))))).... ( -29.40) >DroSec_CAF1 17204 108 - 1 UGUUAUUUAGGCGUGUUCAAGGUUCCGCCUCGAACGCCACAUGGAGUGGAA------UGUAACUUGCAAGUUCGCCAUGAAUACGCUAA-UAAUAAUAUAUAUAUUUGAUAA--CAU (((((((..(((((((((((((.....)))(((((.((((.....))))..------(((.....))).)))))...))))))))))..-.)))))))..............--... ( -30.10) >DroSim_CAF1 20985 108 - 1 UGUUAUUUAGGCGUGUUCGAGGUUCCGCCUCGAACGCCACAUGGAGUGGAA------UGUAACUUGCAAGUUCGCCAUGAAUACGCUAA-UAAUAAUAUAUAUAUUUGAUAA--CAU (((((((..(((((((((((((.....))))((((.((((.....))))..------(((.....))).)))).....)))))))))..-.)))))))..............--... ( -32.70) >DroEre_CAF1 35156 106 - 1 UGUUAUUUAGGCGUGGCCUAGGUUCCGCCUCGAAUGCCACAUGGAGUGGAA------UGUAACUUCCAAGUUCGCCAUGAAUGUGCUUA-UAAUAAUGU--GUUUUUGACAA--AAU (((((((.(((((..((....))..))))).))..(((((((((..(((((------......)))))......)))))..)).))...-.........--.....))))).--... ( -27.00) >DroYak_CAF1 35471 112 - 1 UGUUAUUCAGGCGUGUUCGAGGUUCCGCCUCGAACGCCCAAUGGAGUGGAAUGUGAAUGUAACUUCCAAGUUCGUCAUGAAAAUGUUUA-UAUUAAUAU--GUUUUUGAUAA--UAU .........((.((((((((((.....))))))))))))...(((.(((((.((.......)))))))..)))((((.(((.(((((..-....)))))--.))).))))..--... ( -32.80) >consensus UGUUAUUUAGGCGUGUUCGAGGUUCCGCCUCGAACGCCACAUGGAGUGGAA______UGUAACUUGCAAGUUCGCCAUGAAUACGCUAA_UAAUAAUAUAUAUAUUUGAUAA__CAU ...(((((((((((((((((((.....)))))))))).((.((((((..............)))).)).))..))).)))))).................................. (-17.22 = -18.51 + 1.29)

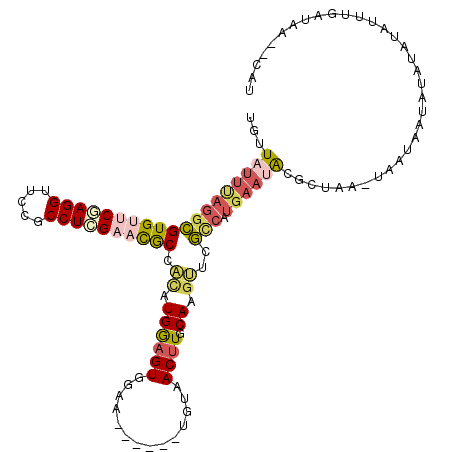
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:05 2006