| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,541,235 – 21,541,340 |
| Length | 105 |
| Max. P | 0.705616 |

| Location | 21,541,235 – 21,541,340 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -24.00 |
| Energy contribution | -25.86 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
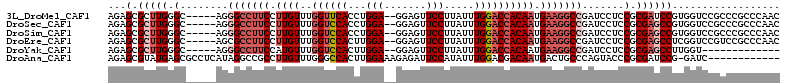
>3L_DroMel_CAF1 21541235 105 + 23771897 AGAGCGCUUGGGC-----AGGGCCUUCCUUGUUUGGUUCACCUGGA--GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAUCCGUGGUCCGCCCGCCCAAC .......((((((-----..(((((((.((((..((((((....((--((....))))...)))))))))).)))))))......(((((...))))).......)))))). ( -43.60) >DroSec_CAF1 12785 105 + 1 AGAGCGCUUGGGC-----AGGGCCUUCCUUGUUUGGUCCACCUGGA--GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAGCCGUGGUCCGCCCGCCCAAC .......((((((-----..(((((((.((((..((((((....((--((....))))...)))))))))).)))))))........(((..(....)..)))..)))))). ( -48.10) >DroSim_CAF1 13658 105 + 1 AGAGCGCUUGGGC-----AGGGCCUUCCUUGUUUGGUCCACCUGGA--GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAGCCGUGGUCCGCCCGCCCAAC .......((((((-----..(((((((.((((..((((((....((--((....))))...)))))))))).)))))))........(((..(....)..)))..)))))). ( -48.10) >DroEre_CAF1 30483 105 + 1 AGAGCGCUUGGGC-----AGCGCCUUCCUUGUUUGGUCCACUUGGA--GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAGCCUCGGUCCGUCCGCCCAAC .(.(((..(((((-----...((((((.((((..((((((....((--((....))))...)))))))))).))))))(((..(((...)))..))))))))..))).)... ( -43.50) >DroYak_CAF1 30863 92 + 1 AGAGCGCUUGGGC-----AGGGCCUUCCAUGUUUGGUCCACUUGGA--GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAGCCUUGGU------------- .(((.(((((((.-----(((((((((.((...(((((((....((--((....))))...)))))))..))))))))....))))).))))))))...------------- ( -39.30) >DroAna_CAF1 14332 99 + 1 AGAGCGUAUGAGCGCCUCAUAGGCCGCCUUGUUUGGGCCACUUGGAAAGAGAUUCCAUAUUUGGACGACAAUGACUGCCCAGUACCCGCGAUCCG-GAUC------------ .(.((.((((((...)))))).)))((((.....))))....(((((.....))))).(((((((((......(((....))).....)).))))-))).------------ ( -26.80) >consensus AGAGCGCUUGGGC_____AGGGCCUUCCUUGUUUGGUCCACCUGGA__GGAGUUCCUUAUUUGGACCACAAUGAAGGCCGAUCCUCCGCGAGCCGUGGUCCGCCCGCCCAAC ...(.(((((.(........(((((((.((((..((((((...(((.......))).....)))))))))).))))))).......).)))))).................. (-24.00 = -25.86 + 1.86)


| Location | 21,541,235 – 21,541,340 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -23.90 |
| Energy contribution | -27.52 |
| Covariance contribution | 3.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21541235 105 - 23771897 GUUGGGCGGGCGGACCACGGAUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC--UCCAGGUGAACCAAACAAGGAAGGCCCU-----GCCCAAGCGCUCU .(((((((((((((((((((.....(((((.....))))).)))))))))...........(((--(...((....)).....))))..)))).-----))))))....... ( -43.10) >DroSec_CAF1 12785 105 - 1 GUUGGGCGGGCGGACCACGGCUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC--UCCAGGUGGACCAAACAAGGAAGGCCCU-----GCCCAAGCGCUCU .(((((((((((..(....)..)))........(((((((.((((((((((....(((....))--).....))))))..)))).)))))))))-----))))))....... ( -49.60) >DroSim_CAF1 13658 105 - 1 GUUGGGCGGGCGGACCACGGCUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC--UCCAGGUGGACCAAACAAGGAAGGCCCU-----GCCCAAGCGCUCU .(((((((((((..(....)..)))........(((((((.((((((((((....(((....))--).....))))))..)))).)))))))))-----))))))....... ( -49.60) >DroEre_CAF1 30483 105 - 1 GUUGGGCGGACGGACCGAGGCUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC--UCCAAGUGGACCAAACAAGGAAGGCGCU-----GCCCAAGCGCUCU .((((((((.((..((((..(((...)))..))))(((((.((((((((((....(((....))--).....))))))..)))).)))))))))-----))))))....... ( -45.30) >DroYak_CAF1 30863 92 - 1 -------------ACCAAGGCUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC--UCCAAGUGGACCAAACAUGGAAGGCCCU-----GCCCAAGCGCUCU -------------.....((..(((...((...(((((((....(((((((....(((....))--).....)))))))......)))))))..-----.))...)))..)) ( -35.60) >DroAna_CAF1 14332 99 - 1 ------------GAUC-CGGAUCGCGGGUACUGGGCAGUCAUUGUCGUCCAAAUAUGGAAUCUCUUUCCAAGUGGCCCAAACAAGGCGGCCUAUGAGGCGCUCAUACGCUCU ------------...(-((.....)))(((.(((((.(((.((((.(.(((....(((((.....)))))..))).)...))))))).(((.....)))))))))))..... ( -28.30) >consensus GUUGGGCGGGCGGACCACGGCUCGCGGAGGAUCGGCCUUCAUUGUGGUCCAAAUAAGGAACUCC__UCCAAGUGGACCAAACAAGGAAGGCCCU_____GCCCAAGCGCUCU .((((((.......((.(.(....).).))...(((((((.((((((((((.....(((.......)))...))))))..)))).))))))).......))))))....... (-23.90 = -27.52 + 3.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:03 2006