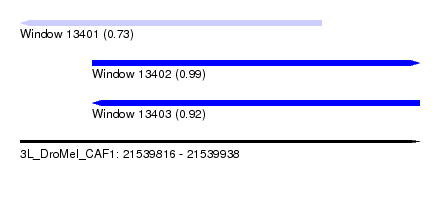
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,539,816 – 21,539,938 |
| Length | 122 |
| Max. P | 0.992050 |

| Location | 21,539,816 – 21,539,908 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21539816 92 - 23771897 ---------CUCCAUUAUUAAUUGUGCGAUUGGAAACUCU-GGUUUUCAAUCACAUGAGUUUCAGAACCUUAGCCCAUUGUCUGCCAUUGCGA------------------GACAUGAAU ---------............(..((.((((((((((...-.)))))))))).))..).................(((.((((((....)).)------------------))))))... ( -21.60) >DroPse_CAF1 40101 111 - 1 ---------CUUUAUUAUUAAUUGUUCGAUUGGAAACUCUUAGUUUUCAAUCACAUGAAACUCAGAAUUAUACUCCAUUGUUUGCUAUUGCUAUUGCUAUUGCUUUUGCCCGACAUGAAU ---------..................((((((((((.....)))))))))).((((.....(((((....((......))..((....((....))....))))))).....))))... ( -18.40) >DroEre_CAF1 29101 92 - 1 ---------CUCCAUUAUUAAUUGUGCGAUUGGAAACUCU-GGUUUUCAAUCACAUGAGUUUCAGAACUUUAGCCCAUUGUCUGCCAUUGCGA------------------GACAUGAAU ---------............(..((.((((((((((...-.)))))))))).))..).................(((.((((((....)).)------------------))))))... ( -21.60) >DroYak_CAF1 29439 92 - 1 ---------CUCCAUUAUUAAUUGUGCGAUUGGAAACUCU-GGUUUUCAAUCACAUGAGUUUCAGAACUUUAGCCCAUUGUCUGCCAUUGCGA------------------GACAUGAAU ---------............(..((.((((((((((...-.)))))))))).))..).................(((.((((((....)).)------------------))))))... ( -21.60) >DroAna_CAF1 12740 102 - 1 UGUAUUGCUGUCUAUUAUUAAUUGUGCGAUUGGAAACUCUUGGUUUUCAAUCACAUGAAAUUCAGAAUUUUACUCCAUUGUUUGUUAUUGCCA------------------GACAUGAAU .(((...(((...(((.....(..((.((((((((((.....)))))))))).))..)))).))).....)))..(((.(((((.......))------------------))))))... ( -20.40) >DroPer_CAF1 49209 111 - 1 ---------CUUUAUUAUUAAUUGUUCGAUUGGAAACUCUUAGUUUUCAAUCACAUGAAACUCAGAAUUAUACUCCAUUGUUUGCUAUUGCUAUUGCUAUUGCUUUUGCCCGACAUGAAU ---------..................((((((((((.....)))))))))).((((.....(((((....((......))..((....((....))....))))))).....))))... ( -18.40) >consensus _________CUCCAUUAUUAAUUGUGCGAUUGGAAACUCU_GGUUUUCAAUCACAUGAAAUUCAGAACUUUACCCCAUUGUCUGCCAUUGCGA__________________GACAUGAAU .....................(..((.((((((((((.....)))))))))).))..).........................((....))............................. (-13.90 = -14.10 + 0.20)


| Location | 21,539,838 – 21,539,938 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 97.81 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21539838 100 + 23771897 CAAUGGGCUAAGGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAAG-CCAUGGCAAAAUAAAUAAAUAAA ......((((.((((......(((.((((((((.((((....)))).))))))))((........))))).....))-)).))))................ ( -21.00) >DroSec_CAF1 11321 100 + 1 CAAUGGGCUAAGGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAAC-CCAUGGCAAAAUAAAUAAAUAAA ......((((.((((......(((.((((((((.((((....)))).))))))))((........)))))....)))-)..))))................ ( -18.80) >DroSim_CAF1 10446 99 + 1 CAAUG-GCUAAGGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAAC-CCAUGGCAAAAUAAAUAAAUAAA .....-((((.((((......(((.((((((((.((((....)))).))))))))((........)))))....)))-)..))))................ ( -18.80) >DroEre_CAF1 29123 100 + 1 CAAUGGGCUAAAGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAAC-CCAUGGCAAAAUAAAUAAAUAAA ..(((((......((((........((((((((.((((....)))).))))))))((........)).))))....)-))))................... ( -18.30) >DroYak_CAF1 29461 101 + 1 CAAUGGGCUAAAGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAACGCCACGGCAAAAUAAAUAAAUAAA .....(((.....((((........((((((((.((((....)))).))))))))((........)).)))).....)))..................... ( -17.40) >consensus CAAUGGGCUAAGGUUCUGAAACUCAUGUGAUUGAAAACCAGAGUUUCCAAUCGCACAAUUAAUAAUGGAGAAAAAAC_CCAUGGCAAAAUAAAUAAAUAAA ......((((.((........(((.((((((((.((((....)))).))))))))((........)))))........)).))))................ (-16.33 = -16.93 + 0.60)

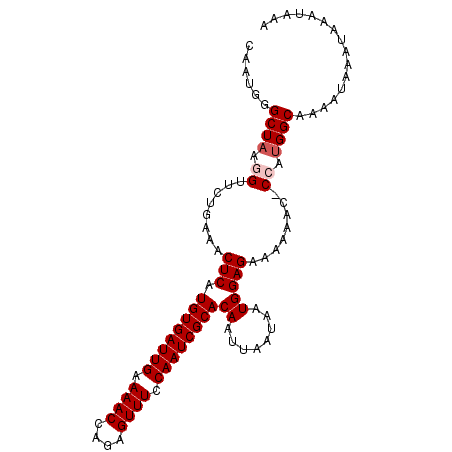
| Location | 21,539,838 – 21,539,938 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 97.81 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21539838 100 - 23771897 UUUAUUUAUUUAUUUUGCCAUGG-CUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACCUUAGCCCAUUG .....................((-((...((((..........(..((.((((((((((....)))))))))).))..).....))))....))))..... ( -20.36) >DroSec_CAF1 11321 100 - 1 UUUAUUUAUUUAUUUUGCCAUGG-GUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACCUUAGCCCAUUG ...................((((-(((..((((..........(..((.((((((((((....)))))))))).))..).....))))....))))))).. ( -23.26) >DroSim_CAF1 10446 99 - 1 UUUAUUUAUUUAUUUUGCCAUGG-GUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACCUUAGC-CAUUG ................((...((-(((((..(((...........(((.((((((((((....)))))))))).))))))....)))))))..))-..... ( -21.70) >DroEre_CAF1 29123 100 - 1 UUUAUUUAUUUAUUUUGCCAUGG-GUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACUUUAGCCCAUUG ...................((((-(((..((((..........(..((.((((((((((....)))))))))).))..).....))))....))))))).. ( -23.26) >DroYak_CAF1 29461 101 - 1 UUUAUUUAUUUAUUUUGCCGUGGCGUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACUUUAGCCCAUUG ...................(.(((.....................(((.((((((((((....)))))))))).)))(((((....)))))..)))).... ( -20.50) >consensus UUUAUUUAUUUAUUUUGCCAUGG_GUUUUUUCUCCAUUAUUAAUUGUGCGAUUGGAAACUCUGGUUUUCAAUCACAUGAGUUUCAGAACCUUAGCCCAUUG ............(((((..((((.(......).))))......(..((.((((((((((....)))))))))).))..)....)))))............. (-17.54 = -17.58 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:01 2006