
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,535,521 – 21,535,692 |
| Length | 171 |
| Max. P | 0.747304 |

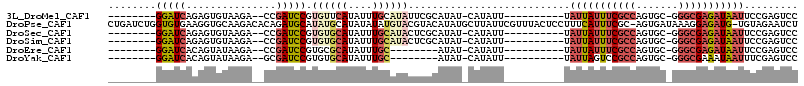
| Location | 21,535,521 – 21,535,614 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -14.60 |
| Energy contribution | -16.16 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21535521 93 + 23771897 --------GGAUCAGAGUGUAAGA--CCGAUCCGUGUUCAUAUUUGCAUAUUCGCAUAU-CAUAUU----------UAUUAUUUCGCCAGUGC-GGGCGAGAUAAUUCCGAGUCC --------(((((.(.((.....)--)))))))(..(((.....(((......)))...-......----------.(((((((((((.....-.)))))))))))...)))..) ( -23.00) >DroPse_CAF1 34519 113 + 1 CUGAUCUGGUGUGAAGGUGCAAGACACAGAUGCAUAUGCAUAUAUAUGUACGUACAUAUGCUUAUUCGUUUACUCCUUUCAUUUCGC-AGUGAUAAAGGAGAUG-UGUAGAAUCU ...(((((((.((......))..)).)))))((((((((((((.(((....))).))))))...........((((((((((.....-.))))..)))))))))-)))....... ( -26.70) >DroSec_CAF1 6917 93 + 1 --------GGAUCAGAGUGUAAGA--CCGAUCCGUGUGCAUAUUUGCAUACUCGCAUAU-CAUAUU----------UAUUAUUUCGCCAGUGC-GGGCGAGAUAAUUCCGAGUCC --------(((((.(.((.....)--)))))))(((((((....)))))))........-......----------.(((((((((((.....-.)))))))))))......... ( -29.10) >DroSim_CAF1 5928 93 + 1 --------GGAUCAGAGUGUAAGA--CCGAUCCGUGUGCAUAUUUGCAUACUCGCAUAU-CAUAUU----------UAUUAUUUCGCCAGUGC-GGGCGAGAUAAUUCCGAGUCC --------(((((.(.((.....)--)))))))(((((((....)))))))........-......----------.(((((((((((.....-.)))))))))))......... ( -29.10) >DroEre_CAF1 24808 85 + 1 --------GGAUCACAGUAUAAGA--CCGAUCCGUGCGCAUAUUUGC--------AUAU-CAUAUU----------UAUUAUUUCGCCAGUGC-GGGCGAGAUAAUUCCGAGUCC --------(((((...........--..)))))(((((......)))--------))..-......----------.(((((((((((.....-.)))))))))))......... ( -22.82) >DroYak_CAF1 25151 85 + 1 --------GGAUCACAGUAUAAGA--GCGAUCCGUGUGCAUAUUUGC--------AUAU-CAUAUU----------UAUUAGUCCGCCAGUGC-GGGCGAAAUAAUUUCGAGUCC --------(((((.(.........--).)))))(((((((....)))--------))))-..((((----------(....((((((....))-)))).)))))........... ( -24.50) >consensus ________GGAUCAGAGUGUAAGA__CCGAUCCGUGUGCAUAUUUGCAUACUCGCAUAU_CAUAUU__________UAUUAUUUCGCCAGUGC_GGGCGAGAUAAUUCCGAGUCC ........(((((...............))))).((((((....))))))...........................(((((((((((.......)))))))))))......... (-14.60 = -16.16 + 1.56)

| Location | 21,535,551 – 21,535,654 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21535551 103 - 23771897 CAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCC-GCACUGGCGAAAUAAUA----------AAUAUG-AUAUGCGAAUAUGCAAAUA ...(((((((((((((....)).)))))).)))))((............(((((.(((((.-.....))))).))))).----------......-((((....))))))..... ( -25.40) >DroPse_CAF1 34559 103 - 1 ----------UGAACGGAAAUGUUUUAGAGUGGAUUUAUAAGAUUCUACA-CAUCUCCUUUAUCACU-GCGAAAUGAAAGGAGUAAACGAAUAAGCAUAUGUACGUACAUAUAUA ----------((.(((...((((((....(((((.((....)).))))).-...(((((((.((...-..))....))))))).........)))))).....))).))...... ( -16.90) >DroSec_CAF1 6947 103 - 1 CAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCC-GCACUGGCGAAAUAAUA----------AAUAUG-AUAUGCGAGUAUGCAAAUA ...(((((((((((((....)).)))))).)))))(((....(((((.((((((.(((((.-.....))))).))))).----------......-...).))))).)))..... ( -30.60) >DroSim_CAF1 5958 103 - 1 CAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCC-GCACUGGCGAAAUAAUA----------AAUAUG-AUAUGCGAGUAUGCAAAUA ...(((((((((((((....)).)))))).)))))(((....(((((.((((((.(((((.-.....))))).))))).----------......-...).))))).)))..... ( -30.60) >DroEre_CAF1 24838 95 - 1 CACGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCC-GCACUGGCGAAAUAAUA----------AAUAUG-AUAU--------GCAAAUA ..((((((((((((((....)).)))))).)))))).............(((((.(((((.-.....))))).))))).----------......-....--------....... ( -25.20) >DroYak_CAF1 25181 95 - 1 CACGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGAAAUUAUUUCGCCC-GCACUGGCGGACUAAUA----------AAUAUG-AUAU--------GCAAAUA ..((((((((((((((....)).)))))).)))))).............((((.((((((.-.....)))))).)))).----------......-....--------....... ( -24.50) >consensus CAUGUCUGUGGAAGCGGAAAUGCCUUCCAGCGGAUGCAAAGGACUCGGAAUUAUCUCGCCC_GCACUGGCGAAAUAAUA__________AAUAUG_AUAUGCGAGUAUGCAAAUA ..((((((((((((((....)).)))))).)))))).............(((((.(((((.......))))).)))))..................................... (-16.93 = -17.68 + 0.76)
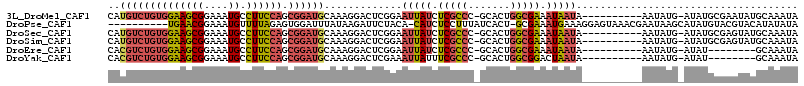


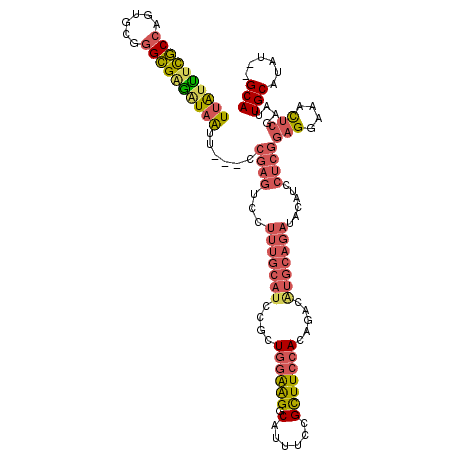
| Location | 21,535,578 – 21,535,692 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -24.73 |
| Energy contribution | -25.98 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21535578 114 + 23771897 UUAUUUCGCCAGUGCGGGCGAGAUAAUU---CCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUGGAAUUGCAUAU--GCA ((((((((((......)))))))))).(---(((((...(((((((...((((.((((......)))).).)))..)))))))......))))))(....)......(((....--))) ( -37.40) >DroSec_CAF1 6974 114 + 1 UUAUUUCGCCAGUGCGGGCGAGAUAAUU---CCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU--GCA .(((((((((......)))))))))(((---(((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))))(((....--))) ( -37.90) >DroSim_CAF1 5985 114 + 1 UUAUUUCGCCAGUGCGGGCGAGAUAAUU---CCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU--GCA .(((((((((......)))))))))(((---(((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))))(((....--))) ( -37.90) >DroEre_CAF1 24857 114 + 1 UUAUUUCGCCAGUGCGGGCGAGAUAAUU---CCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACGUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU--GCA .(((((((((......)))))))))(((---(((((...(((((((...((((.((((......)))).).)))..)))))))......))))(((....)))))))(((....--))) ( -38.20) >DroYak_CAF1 25200 114 + 1 UUAGUCCGCCAGUGCGGGCGAAAUAAUU---UCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACGUGCAGAUGAAUCCUCGGAGGAAACUCGAAUUGCAUAU--GCA ...((((((....)))))).........---.((((((.(((((((...((((.((((......)))).).)))..))))))).))...))))(((....)))....(((....--))) ( -37.30) >DroAna_CAF1 6427 107 + 1 UUGUCUUACAUGCGCGGGUGGGGUUGGUAUGUAAAUUUCUGGAUUUCCGCUCGGGGCCAUUUCCGUUUGCACCUA-----------AUCC-GGGGGGAAAUACGAACUGCAUAUGUGCA ..........((((((.(((.((((.((((.....(..(((((((...((.(((((....)))))...))....)-----------))))-))..)...)))).)))).))).)))))) ( -38.10) >consensus UUAUUUCGCCAGUGCGGGCGAGAUAAUU___CCGAGUCCUUUGCAUCCGCUGGAAGGCAUUUCCGCUUCCACAGACAUGCAGAUACAUCCUCGGAGGAAACUCGAAUUGCAUAU__GCA ((((((((((......))))))))))......((((...(((((((....((((((.(......))))))).....)))))))......))))(((....)))....(((......))) (-24.73 = -25.98 + 1.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:55 2006